Table II.
Approaches for Human Clearance Prediction
| Method | Equation | Data required | Dataset | AFE or APE | <twofold |
|---|---|---|---|---|---|
| Simple allometry (SA) (3,6,11) |
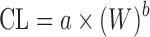 where a and b are the coefficient and exponent of the allometric equation and W is body weight where a and b are the coefficient and exponent of the allometric equation and W is body weight |
CL and body weight in at least two animal species | n = 60 (10) | 3.23 | 81% |
| n = 102 (11) | 2.65 | 54% | |||
| n = 26 (11) | N.A. | 46% | |||
| n = 50 (9) | 1.41 | 53% | |||
| n = 102 (9) | 1.28 | 54% | |||
| n = 103 (42) | N.A. | 53% | |||
| n = 24 (37) | N.A. | 17% | |||
| n = 22 (63) | 2.03 | 68% | |||
| n = 45 (40) | N.A. | 71% | |||
| Allometric scaling of unbound CL (38) | Unbound CL = CL/f u, p | CL, body weight, and plasma unbound (f u, p) in at least two animal species | n = 24 (37) | N.A. | 62% |
| Unbound CL = a × (W)b | n = 12 (12) | 1.79 | 69% | ||
| where f u, p is the unbound fraction in plasma | n = 20 (38) | 2.7 | 55% | ||
| Two term power equation (39) |
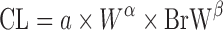 or or 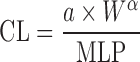
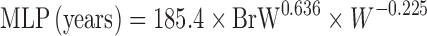 where a is the coefficient and α and β are the exponents of the allometric equation. W and BrW are body weight and brain weight where a is the coefficient and α and β are the exponents of the allometric equation. W and BrW are body weight and brain weight |
CL and body weight in at least three animal species | N.A. | N.A | N.A. |
| Rule of exponents (RoE) (3) | If 0.55 < b <0.70, CL = a × (W)b | CL, body weight, and brain weight in at least two animal species | All categories | ||
| If 0.71 < b < 0.99, CLhuman = a × (MLPanimal × CLanimal)b / MLPhuman | n = 60 (10) | 1.85 | 81% | ||
| If b ≥ 1, CLhuman = a × (BrWanimal × CLanimal)b / 1.53 | n = 102 (11) | 2.25 | 54% | ||
| If b > 1.3, CL will be overpredicted | n = 103 (42) | N.A. | 49% | ||
| If b < 0.55, CL will be underpredicted | n = 24 (37) | N.A. | 37% | ||
| n = 45 (40) | N.A. | 93% | |||
| Allometric scaling for renally and biliarily excreted drugs (7,44) | For renal secreted drugs: 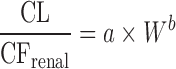
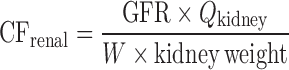
|
CL, body weight, GFR, kidney blood flow, kidney weight, bile flow, and UDPGT activity in at least two animal species | Renal-secreted drugs | ||
For biliary excreted drugs: 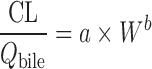 or or 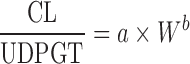
|
n = 10 (43) | N.A. | 70% | ||
| Biliary excreted drugs, n = 8 (44) | N.A. | 50% | |||
| Allometric scaling after normalization by CLint, in vitro (8,45) |
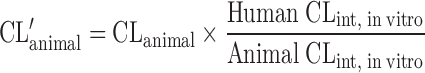
|
CL and body weight in at least two animal species, human and animal microsomal or hepatocyte K m, V max, or concentrations | Extensively metabolized compounds | ||
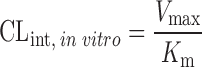 or or 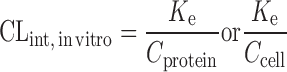
|
n = 10 (8) | N.A. | 80% | ||
| n = 11 (14) | 1.6 | 82% | |||
| Multi-exponential Allometry (MA) (9,48) |
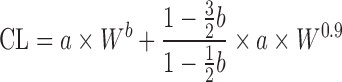 where a and b are the coefficient and exponent of SA where a and b are the coefficient and exponent of SA |
CL and body weight in at least two animal species | All categories | ||
| n = 50 (9) | 1.19 | 76% | |||
| n = 102 (9) | 1.39 | 54% | |||
| n = 45 (48) | N.A. | 89% | |||
| Liver blood flow method (LBF) (11,49) |
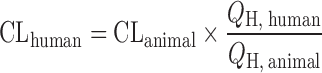
|
Human and animal liver blood flow, animal CL | All categories | Rat 2.57 | Rat, 45% |
| n = 102 (11) | Dog 2.79 | Dog, 50% | |||
| Monkey 1.89 | Monkey, 70% | ||||
| n = 103 (49) | N.A. | Rat, dog, 66% | |||
| N.A. | Monkey, 72% | ||||
| n = 103 (42) | N.A. | Rat, 40% | |||
| N.A. | Dog, 44% | ||||
| N.A | Monkey, 68% | ||||
| Scaling from one or two animal species (11) |
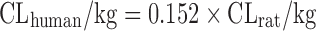
|
CL in at least one species of rat, dog, or monkey | All categories | Rat, 58% | |
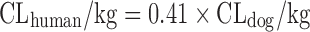
|
Dog, 33% | ||||
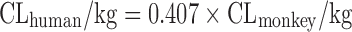
|
n = 26 (11) | N.A. | Monkey, 44% | ||
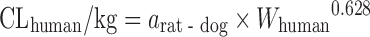
|
Rat–dog, 55% | ||||
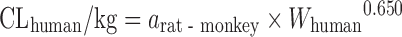
|
Rat–monkey, 78% | ||||
| Vertical allometry (3,6) and f u corrected intercept method (FCIM) (10,40) | Vertical allometry (VA) criteria: ClogP > 2; f u, rat/f u, human > 5; metabolic elimination | ClogP, elimination route, CL, and body weight in at least two animal species, plasma unbound fraction in rats and humans | All categories | ||
FCIM approach: 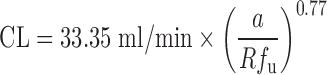 where a is the coefficient of SA and Rf
u is the ratio of unbound fraction in plasma between rats and human where a is the coefficient of SA and Rf
u is the ratio of unbound fraction in plasma between rats and human |
n = 60 (10) | 78 | 90% | ||
| n = 24 (37) | N.A. | 50% | |||
| n = 40 (40) | N.A. | 89% | |||
| In vitro–in vivo extrapolation (IVIVE) using physiological scaling factors (12,23,60) |

|
Drug disappearance rate in human microsomes or hepatocytes | All categories | ||
Well-stirred model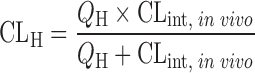
|
n = 33 (13) | 6.17 | 15% | ||
Parallel tube model 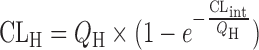
|
n = 22 (63) | 2.01 | 64% | ||
Dispersion model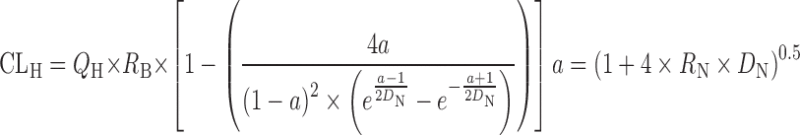
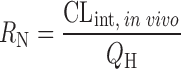 D
N is the dispersion number
D
N is the dispersion number |
n = 7 (12) | 1.95 | 86% | ||
| n = 29 (65) | 2.28 | 79% | |||
| IVIVE using a drug-specific scaling factor (13,60,61) |
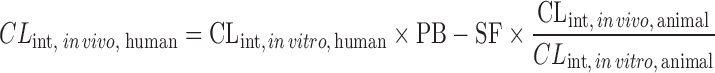
|
Drug disappearance rate in human and animal microsomes or hepatocytes, animal hepatic clearance, the fraction unbound in animal plasma, the animal blood-to-plasma concentration ratio | All categories | ||
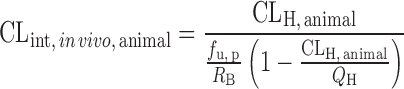
|
n = 33 (13) | 2.33 | 39% | ||
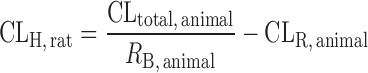
|
n = 13 (60) | 1.57 | 69% | ||
| where PB-SF is the physiologically based scaling factor. The ratio of animal in vivo and in vitro CLint is the drug-specific scaling factor | 1.68 | 77% | |||
| IVIVE using an empirical scaling factor (13,62–64) |
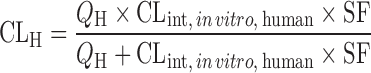 , where SF is the empirical scaling factor, determined by the nonlinear iterative least squares regression between CLint, in vivo and CLint, in vitro
, where SF is the empirical scaling factor, determined by the nonlinear iterative least squares regression between CLint, in vivo and CLint, in vitro
|
Drug disappearance rate in human and rat microsomes or hepatocytes, a dataset of human CLint, in vivo and CLint, in vitro | All categories | ||
| n = 33 (13) | 1.00 | 46% | |||
| n = 22 (63) | 1.64 | 64% | |||
| n = 52 (64) | 1.8–2.2 | 62–76% | |||
| IVIVE with protein binding correction or at the presence of human plasma (67,70) | Well-stirred model 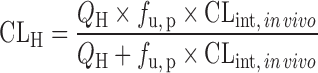
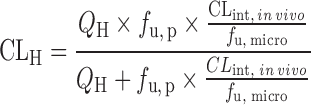
|
Drug disappearance rate in human microsomes or hepatocytes, the fraction unbound in human plasma (f u, p), and liver microsomes (f u, micro), or hepatocytes (f u, hepa) | All categories | ||
Hallifax equation: 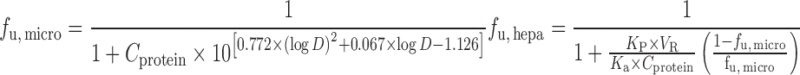
|
n = 7 (12) | 9.28 | 57% | ||
| n = 29 (65) | f u, p 4.39 | 45% | |||
| f u, p, f u, m 2.31 | 62% | ||||
| n = 8 (97) | N.A. | 50% | |||
| n = 7 (67) | N.A. | 86% | |||
| n = 10 (70) | N.A. | 80% | |||
| IVIVE using recombinant P450 enzymes (68) | Relative abundance approach Relative activity approach Relative activity approach |
Intrinsic activity of each recombinant CYP enzymes for each test compound, relative abundance, or activity of CYP enzymes | Benzodiazepines | ||
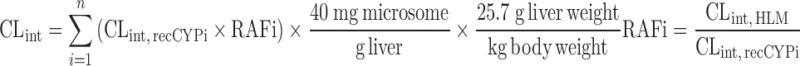
|
n = 5 (69) | N.A. | 80% | ||
| n = 72 (68) | N.A. | 44% | |||
| n = 15 (98) | N.A. | 93% | |||
| Physiologically based approach for renal clearance prediction (34) |
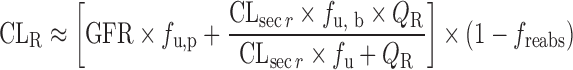
|
GFR, f u, p, CLsecr, and P e | N.A. | N.A. | N.A. |
| Computational approaches: multiple linear regression (MLR) (72), partial least squares (PLS) (73), artificial neural network (ANN) (63,97), and k-nearest neighbors approach (kNN) (74) | Multivariate linear regression analysis 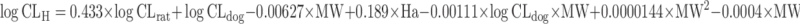
|
Molecular weight and hydrogen bond acceptor number of the test compound, CL in dogs and rats | All categories | ||
| n = 68 (72) | N.A. | 74% | |||
| n = 50 (71) | 1.28 | 78% | |||
| Extensively metabolized compounds n = 22 (63) | 1.81 | 68% |
