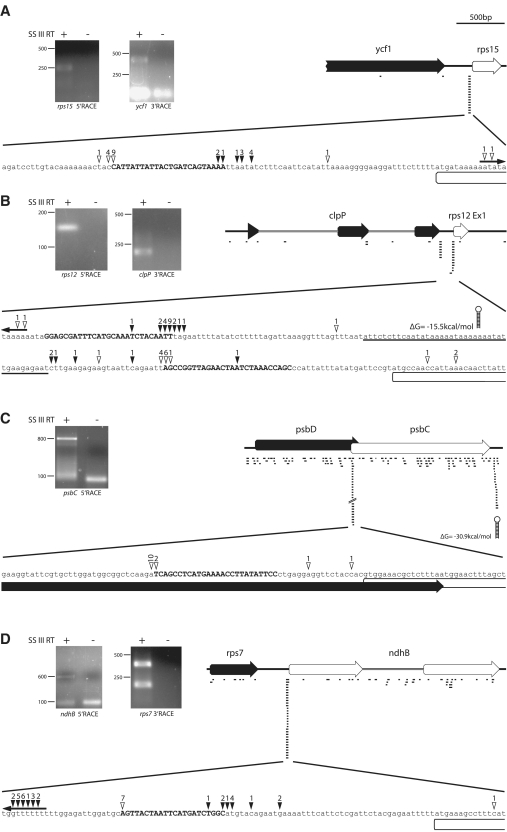
Figure 5.
Mapping of transcript ends in the vicinity of selected sRNAs (A = rps15 5′; B = rps12 5′; C = psbD-psbC; D = rps7-ndhB). Total Arabidopsis RNA was ligated with RNA oligos selectively either at the 3′- or at the 5′-end, reverse-transcribed and amplified by PCR with combinations of gene-specific and oligo-specific primers.The amplification products were separated on an agarose gel (left side of each panel). PCR products were gel-purified and cloned. Clones were selected and sequenced. The last base before the sequence of the RNA oligo corresponds to the end of the original chloroplast RNA ligated. These ends are indicated by open arrowheads (for 5′-RACE experiments) or by filled arrowheads (for 3′-RACE experiments) above a blowup of a sequence stretch covering parts of the intergenic region containing the sRNAs (upper case) at the center. The numbers above the arrowheads point out numbers of clones that correspond to a particular transcript end. In case of 5′-RACE, the numbers refer to independent clones as evidenced by different bar-codes introduced via randomized nucleotides in the ligated 5′-RNA oligo. Number of clones indicating independent ends outside of the blowup region are indicated above outward-facing arrows at the ends of the sequence shown here. All further symbols and numbers are explained in Figure 1.