Figure 3.
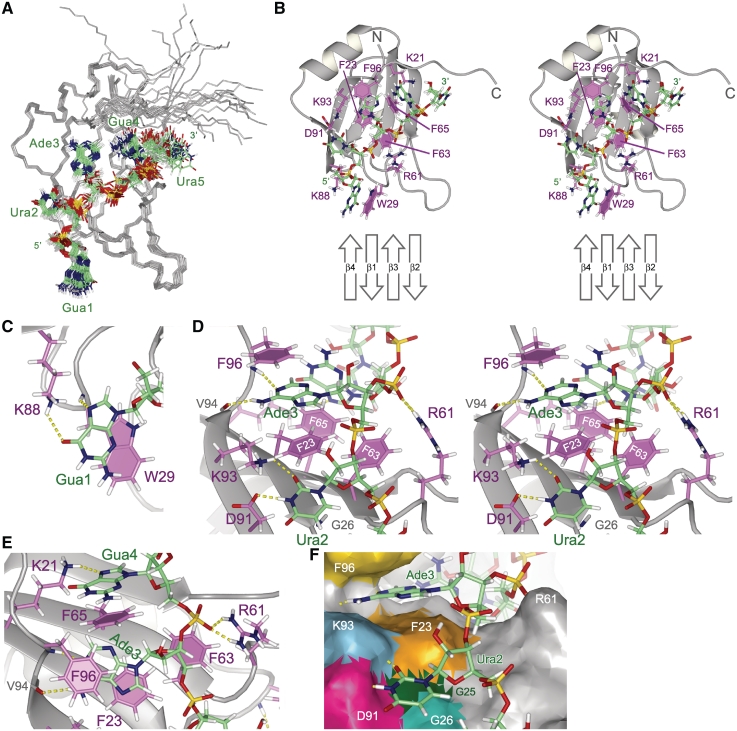
Structure of Msi1 RBD1 in a complex with r(GUAGU) (numb5). (A) Superpositioning of the 20 lowest energy conformers of the RBD1:numb5 complex. The protein backbone (residues 20-103) is colored gray. RNA is shown as a stick model: hydrogen (pale gray), carbon (green), nitrogen (blue), oxygen (red) and phosphorus (yellow). (B) The lowest energy conformer is presented and viewed from the same direction as in (A). Protein side chains contributing to the RNA binding are shown as a stick model: hydrogen (pale gray), carbon (magenta), nitrogen (blue) and oxygen (red); and aromatic rings are filled. RNA is represented as in (A). A schematic drawing of the RNA-binding β-sheet is presented at the bottom. (C) Recognition of Gua1. The Gua1 base stacks onto the W29 indole ring; and hydrogen bonds, Gua1 N7-W29 HN and Gua1 O6- K88 Hζ, are formed. (D) Recognition of Ade3. The Ade3 base is sandwiched between the aromatic rings of F23 and F96; and hydrogen bonds, Ura2 N3-D91 Oδ, Ura2 O2-K93 Hζ, Ade3 H6-V94 O and Ade3 N1-F96 HN, are formed. (E) Recognition of Gua4. The Gua4 base stacks onto the F65 aromatic ring, and Gua4 N7 hydrogen bonds with K21 Hζ. R61 Hη may form a salt bridge with the 5′ phosphate group of Gua4. (F) The Ura2 base stacks in the pocket. The solvent accessible surface of RBD1 is viewed from a similar direction to as in (D). D91, K93, F23 and F63 form a rim, and G25 and G26 form the bottom of the pocket. Hydrogen bonds are indicated by dotted yellow lines. (B) and (D) are stereo diagrams.