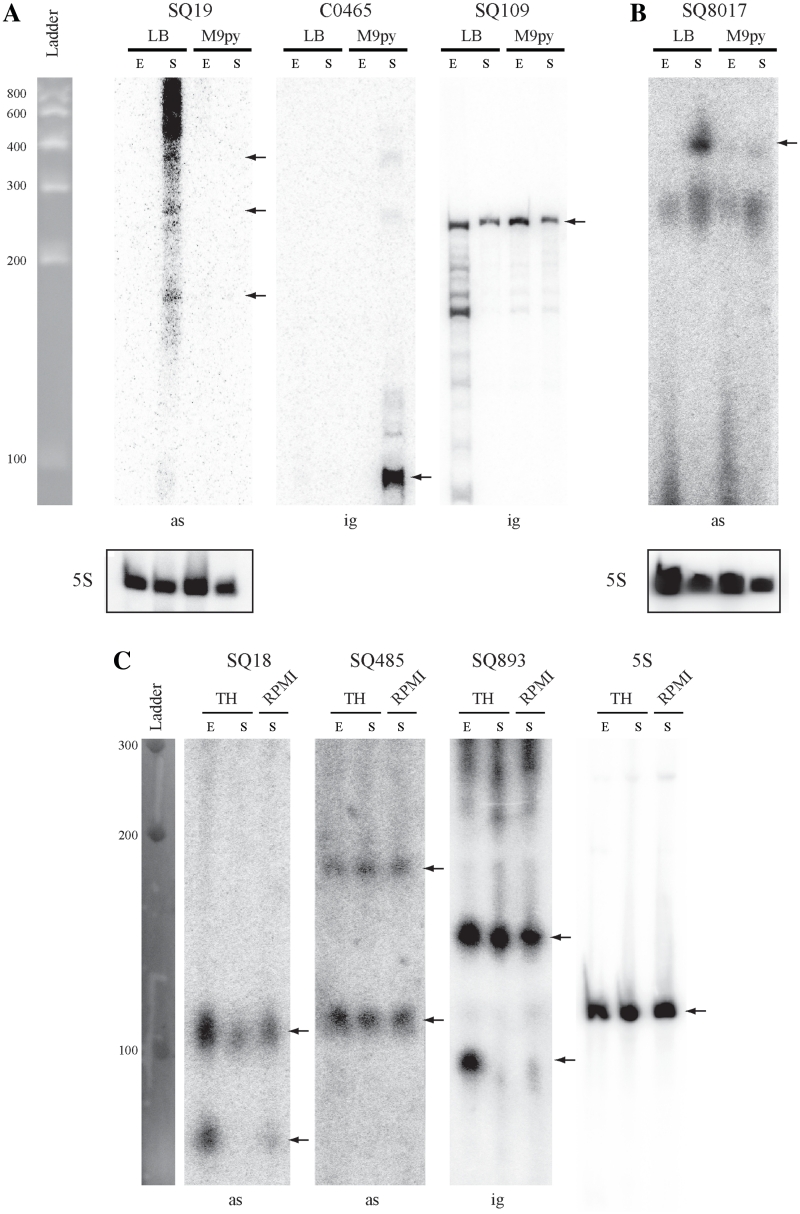
Figure 3.
Northern blot analysis of some sRNAs from E. coli AL862, 536 and S. agalactiae NEM316 strains. Expression analysis of 7 sRNA candidates co-localized with virulence factors (see Table 4) and identified in (A) E. coli AL862, (B) E. coli 536 and (C) S. agalactiae NEM316 strains. Expression was analyzed in two phases of growth (E, exponential; S, late stationary) in LB and M9 + 0.4% pyruvate (M9py) media for E. coli or TH and RPMI1640 + 0.4% glucose media for S. agalactiae. Expression of the constitutively transcribed 5S ribosomal gene was used as loading control. The C0465 sRNA which is expressed only in early stationary phase in E. coli MG1655 strain was used as a negative expression control (46). Notes, ig, sRNA gene located in the IGR; as, sRNA gene located a position antisense to a CDS (asRNA). Black arrows indicated hybridized sRNA molecules.