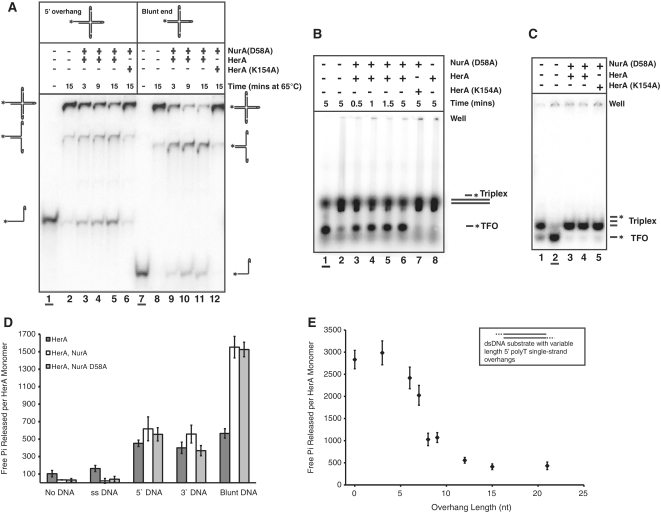
Figure 2.
The nature of the DNA end encountered by the complex affects the outcome of HerA–NurA processing. (A) Holliday junction branch migration, and strand unwinding by the HerA–NurA D58A complex. The 900 nM HerA hexamer and NurA D58A dimer were incubated with the Holliday junction substrate at 65°C for 3, 9 or 15 min. Lanes 1–6: substrate with a single 5′ overhang. Lanes 7–12: blunt-ended substrate. Boiled substrates in lanes 1 and 7 (underlined). Lanes 2 and 8: no protein added (65°C for 15 min). Lanes 6 and 12: HerA K154A–NurA D58A controls. (B) Triplex displacement assay. Lane 1 (underlined), boiled triplex (TFO, triplex forming oligonucleotide); lane 2, no protein added; lanes 3–6, time course of HerA–NurA D58A (230 nM HerA hexamer and NurA D58A dimer); lane 7, HerA K154A–NurA D58A; lane 8, HerA alone. Assays were performed at 50°C using 5 nM triplex DNA. Reactions were resolved on a 1% agarose gel. (C) Triplex displacement control experiment, with the dsDNA shortened to the length of the TFO. Lane 1, no protein added; lane 2 (underlined) boiled sample; lanes 3 and 4 wild-type (230 or 94 nM HerA hexamer and NurA D58A dimer, respectively) HerA–NurA D58A complex after incubation at 50°C for 3 min, respectively; lane 5, HerA K154A–NurA D58A control. Reactions were resolved on a 6% polyacrylamide gel. (D) ATPase assays demonstrating free Pi released per HerA monomer over 20 min (mean value for three independent experiments). Reactions in the absence or presence of DNA for HerA, HerA–NurA and HerA–NurA D58A (40 nM hexamer or dimer in the absence of DNA, or 20 nM for reactions containing both DNA and protein). Error bars depict the standard deviation (SD). (E) HerA ATP turnover versus 5′ overhang length. HerA–NurA complex (20 nM hexamer and dimer) was incubated with dsDNA substrates possessing varying length 5′ polyT overhangs. Data points represent the mean value for five independent experiments.