Figure 4.
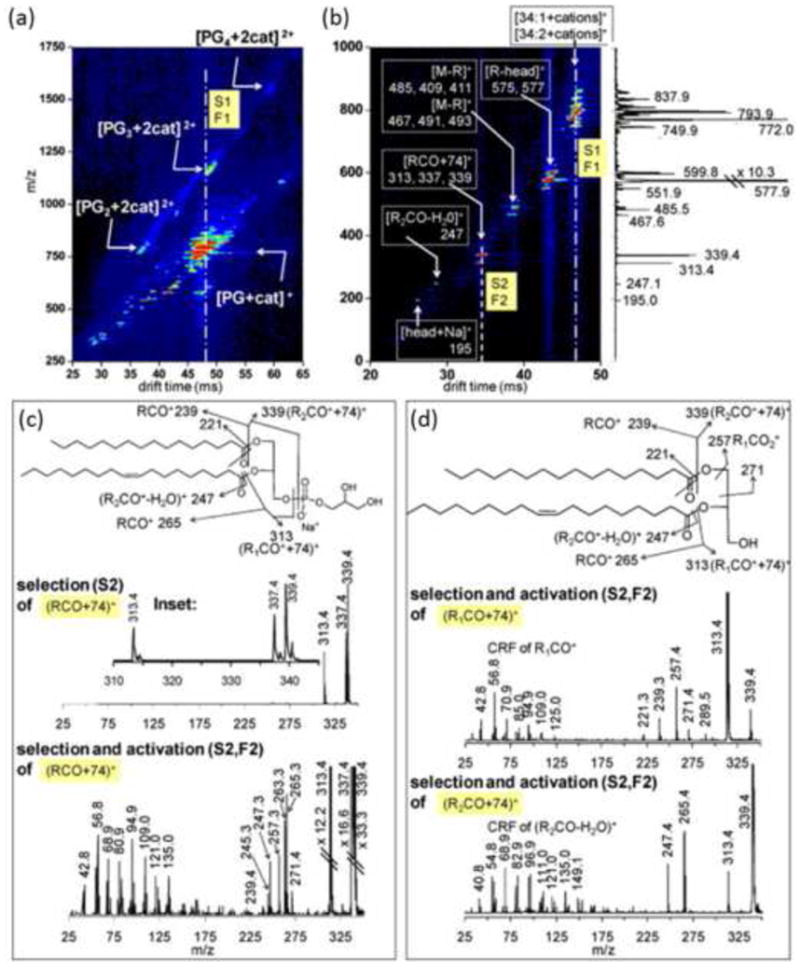
(a) DTIM-MS spectrum of multiply charged multimers of PG lipids demonstrating the ion mobility separation of isobaric lipid ion signals in the 2D IM-MS analysis. (b) DTIM-MS spectrum resulting from tandem ion mobility selection and fragmentation (IM/IM-MS) of the drift time region highlighted by the dotted line in panel (a), which pertains to predominately singly charged monomer ions. (c, top) Mass spectrum resulting from selection of the drift time window highlighted in panel (b). Panels (c, bottom) and (d) represent a second stage of ion fragmentation whereby specific fragment ions generated from the first stage of tandem IM are activated (IM/IM/IM-MS). This is analogous to a higher order tandem MS experiment (e.g., MS3) except that a complete IM-MS spectrum is generated for each stage of ion activation/fragmentation, providing additional structural information regarding fragment ions. Reprinted from Int. J. Mass Spectrom. 287(1-3), Trimpin, S.; Tan, B.; Bohrer, B.C.; O'Dell, D.K.; Merenbloom, S.I.; Pazos, M. X.; Clemmer, D. E.; Walker, J. M.; Profiling of Phospholipids and Related Lipid Structures Using Multidimensional Ion Mobility Spectrometry-Mass Spectrometry, 58-69, Copyright (2008), with permission from Elsevier B.V.
