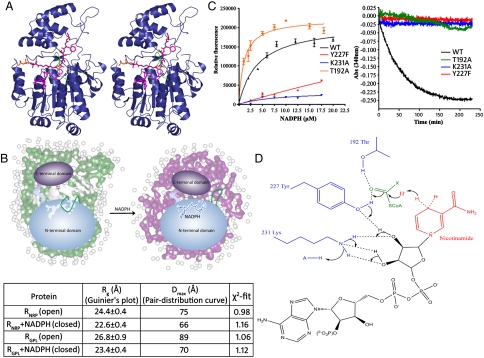
Fig. 4.
Dissecting the two-step 4e- reduction mechanism: (A) Stereoimage shows the cofactor NADPH (magenta) and substrate valeryl-FTAA-ppant (red) modeled in the R domain. A movement of the loop, S153-F166 (green) would provide a platform for binding of the NADPH followed by valeryl-FTAA-ppant. The extrusion of the aldehyde formed after first reduction might facilitate the exchange of oxidized cofactor NADP+ for NADPH. (B) Structural parameters determined for RGPL and RNRP along with NADPH using Guinier approximation and indirect Fourier transformation of SAXS data. Also shown is an illustrative model demonstrating loop movement along with compaction of N- and C-terminal domains in open and closed conformations. (C) NADPH binding to RGPL and Y227F, K231A, and T192A mutants as determined by intrinsic NADPH fluorescence (Left) and analysis of NADPH consumption in spectrophotometric assays of these mutants and WT RGPL using valeryl-FTA-Alaninal substrate (Right). (WT RGPL Kd for NADPH was calculated as 4.64 ± 0.94 μM.) (D) Proposed mechanism of R domain-catalyzed reduction of lipopeptidyl thioester substrate.