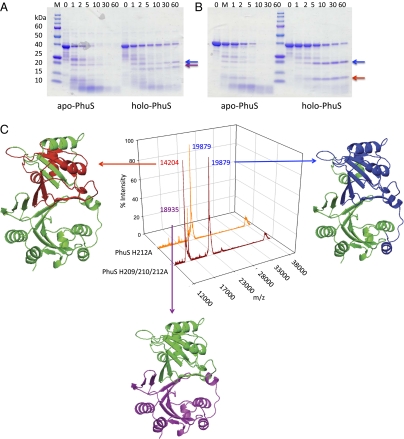
Fig. 2.
Limited proteolysis and MALDI-MS of the wild-type and mutant apo- and holo-PhuS proteins. SDS/PAGE of (A) wild-type apo- and holo-PhuS and (B) apo- and holo-PhuS H209/210/212A. Samples (10 μL) at various time intervals as marked were separated on 8–20% Tris•HCl gradient gels. (C) MALDI-MS spectra of holo-PhuS H212A and holo-PhuS H209/210/212A after 60-min limited proteolysis. The major peptide fragments are highlighted and color coded according to the homology model analysis. The peak at ≈39 kDa corresponds to intact PhuS. The homology model of PhuS was generated from the apo-ChuS crystal structure (Protein Data Bank file 1U9T) using Swiss-Model as previously described (8).