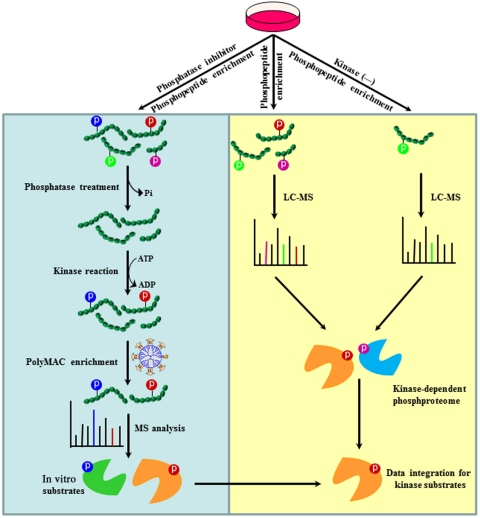
Fig. 1.
Workflow for the KALIP strategy to identify kinase candidates. It involves in vitro kinase reaction and in vivo phosphoproteome. In the in vitro kinase reaction, the critical step is the generation of substrate peptides, which are directly isolated from cell lysate through affinity purification and dephosphorylation. After the kinase reaction, phosphopeptides are further enriched and analyzed by mass spectrometry for sequencing and site identification. In in vivo phosphoproteome, phosphopeptides are enriched from two cell lines (+/- kinase) and analyzed by mass spectrometry. Kinase-modulated phosphorylation events are identified by comparing two phosphoproteomes (qualitatively or quantitatively). Phosphopeptides present within both datasets from in vitro kinase reaction and in vivo phosphoproteomics represent candidate proteins with the highest probability of being genuine Syk substrates. LC-MS, liquid chromatography-mass spectrometry.