Figure 2.
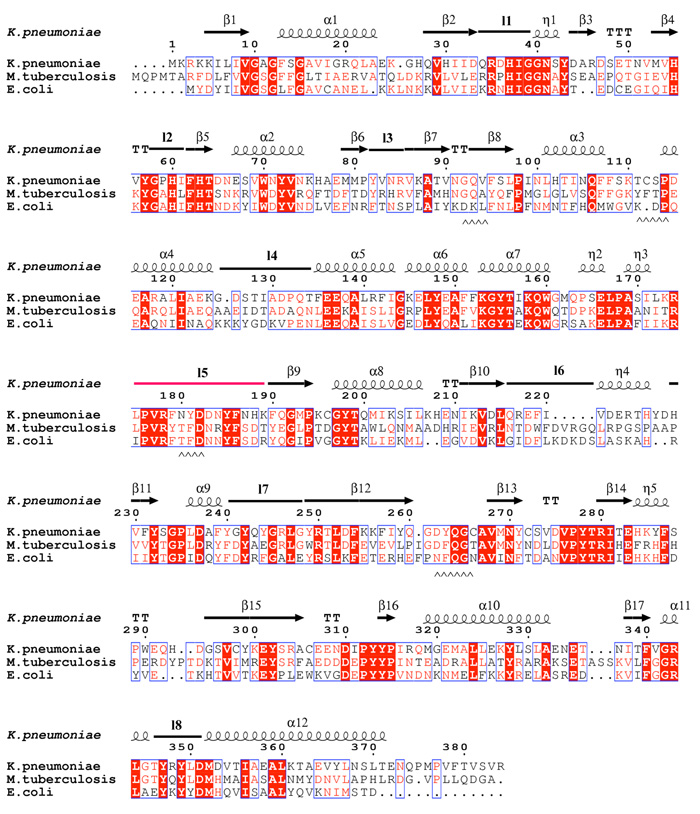
a Sequence alignment of K. pneumoniae with the M. tuberculosis and E. coli mutase enzymes. The alignment is guided by structure and the secondary structure elements for the K. pneumoniae enzyme are shown. The absolutely conserved residues are boxed in red, conservatively substituted residues are shown in red text. Stretches of conservation are shown boxed. The flexible loop 5 discussed in the text is highlighted in the figure in pink. The level of identity is approximately 40%. Residues at the dimer interface are underlined with ^^ symbols.
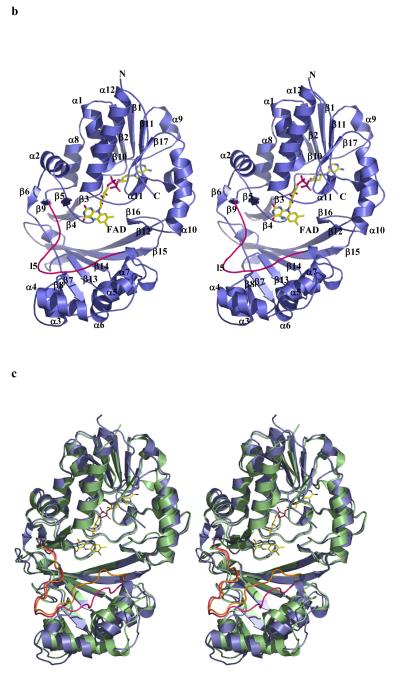
b The monomer structure of the K. pneumoniae enzymes, secondary structure elements are labelled (consistent with Figure 2a). Loop 5 is shown highlighted in pink. FAD sits facing into a large open cleft.
c Superposition of K. pneumoniae (coloured as Figure 2b) enzyme with E. coli enzyme shown in green. The flexible loop in E. coli is coloured orange.
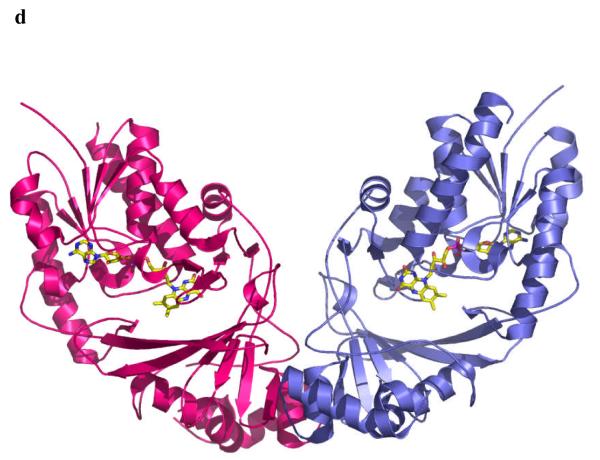
d The dimer structure of the M. tuberculosis enzyme. The dimeric arrangement is conserved in all structures of the enzyme from different organisms.
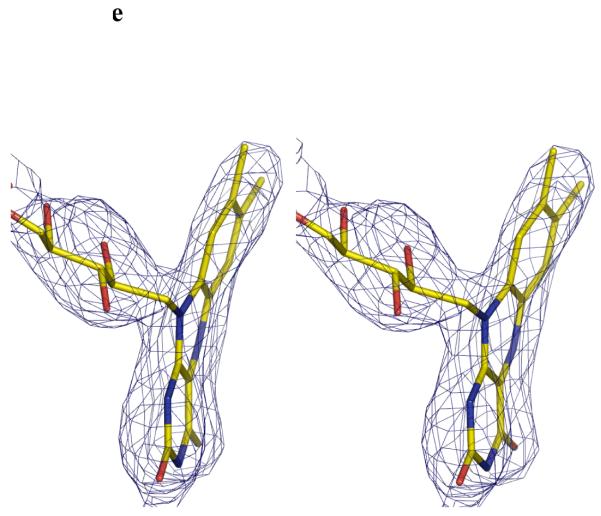
e Stereo image FADH− isoalloxazine ring. The unbiased Fo-Fc map contoured at 3σ for FADH−. FADH− is shown in stick representation. The curvature is towards the si-face. The N5 atom is moved towards the protein and out of the cleft.