Abstract
Introduction
Cytokines play a critical role in regulating the immune system. In the tumor microenvironment, they influence survival, proliferation, differentiation, and movement of both tumor and stromal cells, and regulate tumor interactions with the extracellular matrix. Given these biologic properties, there is reason to hypothesize that cytokine activity influences the pathogenesis of non-Hodgkin lymphoma (NHL).
Methods
We investigated the effect of genetic variation in cytokine genes on NHL prognosis and survival by evaluating genetic variation in individual SNPs as well as the combined effect of multiple deleterious genotypes. Survival information from 496 female incident NHL cases diagnosed during 1996–2000 in Connecticut were abstracted from Connecticut Tumor Registry in 2008. Survival analyses were conducted by comparing Kaplan-Meier curves and hazard ratios (HR) were computed using Cox proportional hazard models adjusting for demographic and tumor characteristics for genes that were suggested by previous studies to be associated with NHL survival.
Results
We found that the variant IL6 genotype is significantly associated (HR=0.42; 95%CI: 0.23–0.77) with a decreased risk of death, as well as relapse and secondary cancer occurrence, among those with NHL. We also found that risk of death, relapse, and secondary cancers varied by specific SNPs for the follicular, DLBCL, and CLL/SLL histologic types. We identified combinations of polymorphisms whose combined deleterious effect significantly alter overall NHL survival and disease-free survival.
Conclusion
Our study provides evidence that the identification of genetic polymorphisms in cytokine genes may help improve the prediction of NHL survival and prognosis.
Keywords: Non-Hodgkin lymphoma, Cytokines, Single nucleotide polymorphisms, Survival
Introduction
Although the median survival for non-Hodgkin lymphoma (NHL) is approximately 10 years, the course of the disease is highly variable, progressing slowly for indolent tumors and very rapidly for aggressive tumors [1]. Studies have shown that NHL survival patterns vary by subtype [2,3], suggesting different prognostic risk factors for NHL histological subtypes. Established adverse prognostic factors for NHL include older age at diagnosis, higher tumor stage, poor performance score, extranodal involvement and above-normal lactate dehydrogenase [4]. The investigation of the impact of genetic variation in cytokines on NHL pathogenesis and prognosis is of interest as cytokines are secreted proteins that play a critical role in regulating the immune system; they control lymphoid cell development and differentiation, and regulate the balance between the T-helper immune responses, as well as proliferation, differentiation, and the movement and communication between tumor and stromal cells [5]. Given these biologic properties, investigation of the association between variation in cytokine gene expression and the progression of NHL is important in light of the increasing incidence of NHL observed in recent decades [6].
Since the expression of T-helper cytokines can be altered by germ-line genetic variants, we studied the association between common polymorphisms in Th1 and Th2 cytokine genes and the risk of NHL in this study population previously [7]. We found that SNPs in critical genes, IL4, IL5, IL6, and IL10, were associated with risk of NHL. Subsequent analyses by NHL subtype showed that variants in IL10 and IL5 were significantly associated with an increased risk of B-cell lymphoma, and variants in IL4, IL4R, and IL6 were significantly associated with an altered risk of T-cell lymphoma. The results raised the possibility that a shift in the balance of the Th1/Th2 response caused by genetic variants in key cytokine genes could have important consequences for the pathogenesis of NHL. In addition to investigation of NHL risk, it is possible that cytokine gene expression plays a role in the progression of NHL.
To more broadly test the hypothesis that inherited variability in cytokine and related immune genes affects NHL survival and disease free survival, we evaluated the association of 82 single nucleotide polymorphisms (SNPs) from 40 candidate immune genes (Table 1) in study participants that have been followed post disease diagnosis. We chose candidate SNPs from genes involved in key immune pathways, particularly those related to cytokine regulation and function in a population-based case-control study among women in Connecticut. We hypothesized that the genetic variation in the cytokines and the combined effect of multiple deleterious genotypes affect NHL prognosis and survival, and we examined the associations by NHL subtype.
Table 1.
Candidate genes and SNPs
| Gene | Name | Location | SNP rs number |
|---|---|---|---|
| CARD15 | Caspase recruitment domain family, member 15 | 16q12 | rs2066847, rs2066844 |
| CCR2 | Chemokine, CC motif, receptor 2 | 3p21 | rs1799864 |
| CCR5 | Chemokine, CC motif, receptor 5 | 3p21 | rs1800940, rs1799987, rs1800560 |
| CSF2 | Colony-stimulating factor 2 (granulocyte) | 5q31.1 | rs1469149, rs25882 |
| CTLA4 | Cytotoxic T lymphocyte-associated 4 | 2q33 | rs231775 |
| CX3CR1 | Chemokine, CXC motif | 3p21 | rs3732379 |
| CXCL12 | Chemokine, CXC motif, ligand 12 | 10q11.1 | rs1801157 |
| FCGR2A | Receptor for Fc fragment of IgG, low-affinity IIa (CD32) | 1q21-q23 | rs1801274 |
| ICAM1 | Intercellular adhesion molecule 1 (CD54) | 19p13.3-p13.2 | rs5491 |
| IFNGR1 | Interferon, gamma, receptor 1 | 6q23-q24 | rs3799488 |
| IFNGR2 | Interferon, gamma, receptor 2 | 21q22.11 | rs9808753, rs1059293, rs4986958 |
| IFNG | Interferon | 12q14 | rs1861494, rs2069705 |
| IL10RA | Interleukin 10 receptor, alpha | 11q23.3 | rs9610 |
| IL10 | Interleukin 10 | 1q31-q32 | rs1800871, rs1800872, rs1800896, rs3024509, rs3024496, rs3024491, rs1800890 |
| IL12A | Interleukin 12, alpha | 3q25.33 | rs568408, rs582054 |
| IL12B | Interleukin 12B | 5q33.3 | rs3212227 |
| IL13 | cytotoxic lymphocyte maturation factor 2 | 5q23.3 | rs20541, rs1800925, rs1295686 |
| IL15RA | Interleukin 15 receptor, alpha | 10p15.1 | rs2296135 |
| IL16 | Interleukin 16 | 15q25.1 | rs859, rs11325 |
| IL1A | Interleukin 1-alpha | 2q13 | rs17561, rs1800587, rs2856841 |
| IL1B | Interleukin 1-beta | 2q14 | rs16944, rs1143634, rs1143627 |
| IL1RN | Interleukin 1 receptor antagonist | 2q14.2 | rs454078 |
| IL2 | Interleukin 2 | 4q26-q27 | rs2069762 |
| IL4R | Interleukin 4 receptor | 16p12.1-p11.2 | rs1805011, rs1801275, rs1049631, rs2107356 |
| IL4 | Interleukin 4 | 5q31.1 | rs2243250, rs2243248, rs2070874, rs2243290, rs2243268 |
| IL5 | Interleukin 5 | 5q31.1 | rs2069812, rs2069822, rs2069818, rs2069807 |
| IL6 | Interleukin 6 | 7p15.3 | rs1800795, rs1800797, rs1800796 |
| IL7R | Interleukin 7 receptor (CD127) | 5p13 | rs1494555, rs2228141 |
| IL8RA | Interleukin 8 receptor, alpha | 2q35 | rs2234671 |
| IL8RB | Interleukin 8 receptor, beta | 2q35 | rs1126579, rs2230054, rs1126580 |
| IL8 | Interleukin 8 | 4q12-q13 | rs4073, rs2227307, rs2227306, rs2227538 |
| LEPR | Leptin receptor | 1p31 | rs1805096, rs7602 |
| MIF | Macrophage migration inhibitory factor | 22q11.23 | rs755622, rs9282783 |
| SELE | Selectin E | 1q22-q25 | rs5361 |
| STK11 | Serine/threonine kinase 11 | 19p13.3 | rs9282859, rs741764 |
| TGFB1 | Transforming growth factor, beta 1 | 19q13.2 | rs1800470 |
| TGFBR1 | Transforming growth factor, beta receptor 1 | 9q22 | rs868 |
| VCAM1 | Vascular cell adhesion molecule 1 | 1p32-p31 | rs1041163 |
Materials and methods
Study population
The study population has been described elsewhere [8,9]. In brief, a total of 1,122 potential female NHL cases aged between 21 and 84 years were identified between 1996 and 2000 through the Yale Comprehensive Cancer Center’s Rapid Case Ascertainment Shared Resource (RCA), a component of the Connecticut Tumor Registry (CTR). Among those cases, 167 died before they could be interviewed and 123 were excluded because of doctor refusal, previous diagnosis of cancer (excluding non-melanoma skin cancer), or inability to speak English. Of 832 remaining eligible cases, 601 completed an in-person interview. Pathology slides or tissue blocks were obtained from the hospitals where the cases had been diagnosed. The specimens were reviewed by two independent study pathologists. All NHL cases were classified according to the World Health Organization (WHO) classification system [10,11]. Vital status for these NHL cases was abstracted at the CTR in May-October 2008. Other follow-up information was also abstracted, including date of death, most recent follow-up date, type and date of treatments, dates of relapse and/or secondary cancer, B-symptoms, and tumor stage. There were 250 deaths from any cause and 140 deaths from NHL in the study population. Of the 601 cases, 13 were not able to be identified in the CTR system, 13 were found to have a cancer history prior to diagnosis of NHL, and 79 had genotyping data missing, yielding 496 NHL patients in the final analyses. Of these, 155 had diffuse large B-cell lymphoma (DLBCL); 117 had follicular lymphoma (FL); 57 had chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL); 34 had marginal zone B-cell lymphoma (MZBL); and 37 had T/NK-cell lymphoma (T-cell).
Genotyping
DNA was extracted from blood or buccal cell samples using phenol–chloroform extraction. Genotyping was conducted at the National Cancer Institute Core Genotyping Facility (Advanced Technology Center, Gaithersburg, Maryland, USA) using validated assays on the Taqman (Applied Biosystems, Foster City, California, USA) or MGB Eclipse (Epoch Biosciences, Bothell, Washington, USA) platforms. Sequence data and assay conditions available at: http://snp500cancer.nci.nih.gov [12]. We selected 82 single nucleotide polymorphisms (SNPs) in 40 immune function genes from the designable set of common SNPs (minor allele frequency >5%) genotyped in the Caucasian (CEU) population sample of the HapMap Project (Data Release 20/Phase II, NCBI Build 35 assembly, dpSNPb125) using the software Tagzilla [http://tagzilla.nci.nih.gov/], which implements a tagging algorithm based on the pairwise binning method of Carlson et al. (Table 1) [13]. All 82 SNPs were genotyped in blood-based DNA samples because DNA samples from buccal cell were insufficient for this analysis. When there were multiple transcripts available for genes, the primary transcript was assessed. Duplicate samples from 100 study subjects and 40 replicate samples from each of two blood donors were interspersed throughout the plates used for genotype analysis. The concordance rates for quality control samples were 99–100% for all assays. We observed no significant departure from Hardy-Weinberg equilibrium in the control population for any of the SNPs analyzed (P>0.05).
Statistical analysis
Survival analyses were conducted for both overall survival (OS) and disease-free survival (DFS). In OS analysis, deaths were events and being alive was censoring. In DFS analysis, deaths, relapses and occurrences of secondary cancer were events and otherwise were censorings.
We assumed the Cox proportional hazards (PH) model and estimated hazard ratios (HR) and 95% confidence intervals (95% CI) for the association of each individual genotype with OS and DFS. The homozygote of the most common allele was used as the reference group and coded as 0, and the heterozygote and homozygote variant genotypes were grouped together and coded as 1. The reference groups with a homozygote wild-type genotype were coded as 0, and the heterozygote and homozygote variant genotypes were grouped together to increase power and coded as 1. For most genotypes, the frequency of homozygote variants does not permit analysis. As such, we used the combined approach. Age (continuous), education (high school or less, some college, and college graduate or more), stage (I, II, III, IV, and unknown), B-symptom (yes, no) and initial treatment (none, radiation only, chemotherapy-based regimen, and other) were adjusted as a priori confounder variables. Adjustment for race did not result in material changes of the associations, and thus race was not included in the final model.
A multi-SNP model was fitted for NHL and subtypes respectively. First, the SNPs with p-values <0.15 from above single-SNP analysis were selected and fit in a Cox model adjusting for age, education, stage, B-symptom and initial treatment. Then we ran stepwise (backwards) selection for the Cox model to retain a parsimonious number of SNPs while keeping a priori confounders in the model. We specified a number of criteria for variable deletion including the statistic for a variable to be added must be significant (0.20) and the value at which any variable that does not produce an F statistic significant should be deleted (0.20). Such selection was done for NHL overall as well as each subtype (for both overall survival and disease free survival). A group of four SNPs that concurrently affect NHL survival was identified and the deleterious genotypes were identified from the single-SNP analysis. Kaplan-Meier survival curves were plotted by the number of deleterious genotypes (0, 1, 2, or ≥3) for NHL overall and subtypes. Log-rank statistics were computed to evaluate the difference in survival. The false discovery rate (FDR) method was applied to adjust for multiple comparisons. The FDR provides the expected ratio of erroneous rejections of the null hypothesis compared to the total number of rejected hypotheses. We set FDR=0.2, then calculated p2=FDR*rank/n, where n is total number of comparisons and determined the value to be significant if p2>=p.
Given the parameters of the respective models, we computed the log likelihood of the data for the null and alternative models to determine whether the models including the gene combinations improved predictability. We compared the likelihood ratio to a chi-square test statistic. If, the likelihood ratio was greater than the critical value, we rejected the null hypothesis and concluded that the model including the SNPs made a significant improvement to the model without the SNP data.
Statistical analyses were performed using SAS, version 9.1 (SAS Institute, Cary, NC). The study was approved by the Human Investigation Committee at Yale University and the Connecticut Department of Public Health.
Results
Demographic and tumor characteristics for the 496 NHL cases are presented in Table 2. During follow-up, 211 deaths, 17 recurrence of NHL, and 63 secondary cancers occurred. The mean overall survival was 6.92 years (SD=3.13, range: 0.38–11.68) and the mean disease free survival was 6.61 years (SD=3.20, range: 0.04–11.68).
Table 2.
Demographic and clinical characteristics of NHL cases, Connecticut, 1996–2001
| Number | % | |
|---|---|---|
| Age at diagnosis | ||
| <=45 | 61 | 12.30 |
| 46–55 | 100 | 20.16 |
| 56–65 | 113 | 22.78 |
| 66–75 | 140 | 28.23 |
| >=76 | 82 | 16.53 |
| Race | ||
| White | 475 | 95.77 |
| Black | 16 | 3.23 |
| Other | 5 | 0.80 |
| Education | ||
| High School or Less | 206 | 41.53 |
| Some College | 168 | 33.87 |
| College graduate or more | 122 | 24.60 |
| Family History | ||
| None | 107 | 21.57 |
| Any other cancer | 381 | 76.82 |
| NHL | 8 | 1.61 |
| Stage | ||
| I | 238 | 47.98 |
| II | 61 | 12.30 |
| III | 28 | 5.65 |
| IV | 158 | 31.85 |
| UK | 11 | 2.22 |
| B-symptom | ||
| Yes | 71 | 14.31 |
| No | 425 | 85.69 |
| Initial Treatment | ||
| None | 99 | 19.96 |
| Surgery | 180 | 36.29 |
| Radiation | 39 | 7.86 |
| Chemotherapy | 121 | 24.40 |
| Other | 57 | 11.49 |
Frequencies of SNPs in cytokine genotypes in NHL overall and subtypes are presented in Supplemental Table 1. The non-genotyped rates ranged from 8.5% (IL8RB, ICAM1, FCGR2A) to 56.2% (IL4R).
As shown in Table 3, there was one SNP in IL6 significantly associated (HR=0.42; 95%CI: 0.23–0.77) with the risk of death in NHL cases after adjusting for demographic and clinical risk factors (Table 3). When NHL subtypes were examined separately, one SNP in SELE (rs5361) was found to be significantly associated (HR=0.42; 95%CI: 0.20–0.89) with the risk of death in DLBCL patients; seven SNPs, including CCR2 (rs1799864) (HR=0.27; 05%CI: 0.08–0.86), IFNGR1 (rs3799488) (HR=3.19; 95%CI: 1.09–9.34), IL4R (rs1801275) (HR=2.35; 95%CI: 1.07–5.19), IL8 (rs4073) (HR=2.60; 95%CI: 1.10–6.15), IL8 (rs2227307) (HR=2.57; 95%CI: 1.07–6.17), MIF (rs755622) (HR=2.45; 95%CI: 1.09–5.47) and SELE (rs5361) (HR=0.10; 95%CI:0.02–0.48) were significantly associated with the risk of death in FL patients; and three SNPs in IFNGR2 (rs1059293) (HR=3.02; 95%CI: 1.06–8.16), IL8RB (rs1126579) (HR=3.54; 95%CI: 1.38–9.06), and TGFBR1 (rs868) (HR=0.29; 95%CI: 0.09–0.87) were significantly associated with the risk of death in CLL/SLL patients (Table 3).
Table 3.
Hazard ratios for risk of death associated with SNPs (p<0.15) in cytokine genes for NHL overall and subtypes (n for NHL=496, DLBCL=155, FL=117, CLL/SLL=34)
| Gene | SNP | HR (95% CI) NHL Overall | HR (95% CI) DLBCL | HR (95% CI) FL | HR (95% CI) CLL/SLL |
|---|---|---|---|---|---|
| CCR2 | rs1799864 | 0.27 (0.08–0.86) | |||
| CCR5 | rs1799987 | 0.76 (0.56–1.03) | |||
| CCR5 | rs1800940 | 0.73 (0.53–1.00) | 0.46 (0.19–1.15) | ||
| CSF2 | rs25882 | 2.73 (0.98–7.65) | |||
| CTLA4 | rs231775 | 0.66 (0.37–1.17) | 1.31 (0.61–2.83) | ||
| CX3CR1 | rs3732379 | 1.28 (0.94–1.72) | |||
| FCGR2A | rs1801274 | 1.27 (0.94–1.71) | 1.79 (1.01–3.17) | ||
| IFNGR2 | rs1059293 | 3.02 (1.06–8.16) | |||
| IFNGR1 | rs3799488 | 3.19 (1.09–9.34) | |||
| IL16 | rs11325 | 0.56 (0.28–1.13) | |||
| IL4R | rs1805011 | 1.32 (0.91–1.89) | 1.82 (0.88–3.77) | ||
| IL4R | rs1801275 | 1.28 (0.90–1.84) | 2.35 (1.07–5.19) | ||
| IL4 | rs2243250 | 0.49 (0.19–1.27) | |||
| IL4 | rs2243248 | 0.74 (0.50–1.10) | |||
| IL5 | rs2069812 | 1.25 (0.93–1.69) | |||
| IL6 | rs1800796 | 0.42 (0.23–0.77)a | 0.25 (0.06–1.06) | 0.25 (0.06–1.10) | |
| IL7R | rs2228141 | 1.98 (0.82–4.79) | |||
| IL8 | rs4073 | 2.60 (1.10–6.15) | |||
| IL8 | rs2227307 | 2.57 (1.07–6.17) | |||
| IL8 | rs2227306 | 2.23 (0.98–5.11) | |||
| IL8RA | rs2234671 | 1.74 (0.81–3.84) | |||
| IL8RB | rs1126579 | 3.54 (1.38–9.06)a | |||
| IL12A | rs582054 | 1.51 (0.83–2.76) | |||
| IL10 | rs3024509 | 0.22 (0.04–1.24) | |||
| MIF | rs755622 | 2.45 (1.09–5.47) | |||
| SELE | rs5361 | 0.68 (0.45–1.03) | 0.42 (0.20–0.89)a | 0.10 (0.02–0.48)a | |
| STK11 | rs741764 | 1.52 (0.87–2.64) | |||
| TGFBR1 | rs868 | 1.70 (0.91–3.17) | 0.29 (0.09–0.87)a | ||
| VCAM1 | rs1041163 | 2.92 (0.88–9.71) |
Reference groups were homozygote wild-type carriers. Models adjusted for age (continuous), education (high school or less, some colleges, and college graduate or more), stage (I, II, III, IV and unknown), B-symptom presence (yes, no, and missing) and initial treatment (none, radiation only, chemotherapy-based regimen, and other)
Remained significant at the p=0.05 level after FDR adjustment
One SNP in IL6 (rs1800796) was found to be significantly associated (HR=0.39; 95%CI: 0.22–0.71) with risk of death, relapse or secondary cancer in overall NHL cases (Table 4). When NHL subtypes were examined separately, two SNPs in FCGR2A (rs1801274) (HR=1.78; 95%CI: 1.04–3.04) and in TGFBR1 (rs868) (HR=1.85; 95%CI: 1.02–3.33) were associated with an increased risk of death, relapse or secondary cancer in DLBCL survivors; three SNPs in IFNGR1 (rs3799488) (HR=2.63; 95%CI: 01.00–6.91), IL4R (rs1801275) (HR=2.53; 95%CI: 1.22–5.21), and SELE (rs5361) (HR=0.26; 95%CI: 0.09–0.78) were associated with a risk of death, relapse or secondary cancer in FL survivors; four SNPs in TGFBR1 (rs868) (HR=0.26; 95% CI: 0.09–0.77), IL8RB (rs1126579) (HR=3.33; 95% CI: 1.35–8.20), CSF2 (rs25882) (HR=2.63; 95%CI: 0.99–6.97), and CCR5 (rs1800940) (HR=0.39; 95% CI: 0.16–0.95) was associated with risk of death, relapse or secondary cancer in CLL/SLL survivors (Table 4).
Table 4.
Hazard ratios for risk of death, relapse or secondary cancer occurence associated with SNPs (p<0.15) in cytokine genes for NHL overall and subtypes (n for NHL=496, DLBCL=155, FL=117, CLL/SLL=34)
| Gene | SNP | HR (95% CI) NHL Overall | HR (95% CI) DLBCL | HR (95% CI) FL | HR (95% CI) CLL/SLL |
|---|---|---|---|---|---|
| CARD15 | rs2066844 | 0.31 (0.07–1.38) | |||
| CCR2 | rs1799864 | 0.39 (0.15–1.05) | |||
| CCR5 | rs1800940 | 0.79 (0.60–1.05) | 0.39 (0.16–0.95) | ||
| CSF2 | rs25882 | 2.63 (0.99–6.97) | |||
| CTLA4 | rs231775 | 0.66 (0.39–1.13) | 0.49 (0.21–1.15) | ||
| CX3CR1 | rs3732379 | 1.26 (0.95–1.67) | |||
| FCGR2A | rs1801274 | 1.78 (1.04–3.04) | |||
| IFNGR2 | rs1059293 | 2.51 (0.94–6.74) | |||
| IFNGR1 | rs3799488 | 1.33 (0.97–1.83) | 2.63 (1.00–6.91) | 0.42 (0.14–1.28) | |
| IL4R | rs1805011 | 1.37 (0.98–1.92) | 1.75 (0.88–3.50) | 2.14 (0.99–4.64) | |
| IL4R | rs1801275 | 1.33 (0.99–1.77) | 2.53 (1.22–5.21)a | ||
| IL5 | rs2069812 | 0.69 (0.42–1.14) | |||
| IL5 | rs2069807 | 82.5 (7.15–953.0) | |||
| IL6 | rs1800797 | 1.61 (0.91–2.83) | |||
| IL6 | rs1800796 | 0.39 (0.22–0.71)a | 0.20 (0.05–0.83)a | 0.32 (0.09–1.10) | |
| IL7R | rs2228141 | 1.81 (0.84–3.88) | |||
| IL8 | rs4073 | 2.23 (1.03–4.80) | |||
| IL8 | rs2227307 | 2.15 (0.99–4.65) | |||
| IL8 | rs2227306 | 1.96 (0.93–4.13) | |||
| IL8 | rs2227538 | 108.6 (9.0–1313.2) | |||
| IL8RA | rs2234671 | 1.81 (0.90–3.65) | |||
| IL8RB | rs1126579 | 1.25 (0.92–1.68) | 3.33(1.35–8.20) | ||
| IL12A | rs582054 | 1.56 (0.88–2.96) | |||
| IL12B | rs3212227 | 1.70 (0.87–3.32) | |||
| IL10 | rs3024509 | 2.31 (0.90–5.88) | 0.20 (0.04–1.08) | ||
| IL16 | rs859 | 1.75 (0.87–3.50) | |||
| IL16 | rs11325 | 0.62 (0.33–1.09) | |||
| MIF | rs755622 | 1.86 (0.89–3.89) | |||
| SELE | rs5361 | 0.57 (0.30–1.11) | 0.26 (0.09–0.78)a | ||
| TGFBR1 | rs868 | 1.85 (1.02–3.33) | 0.26 (0.09–0.77)a | ||
| VCAM1 | rs1041163 | 2.48 (0.78–7.92) |
Reference groups were homozygote wild-type carriers. Models adjusted for age (continuous), education (high school or less, some colleges, and college graduate or more), stage (I, II, III, IV and unknown), B-symptom presence (yes, no, and missing) and initial treatment (none, radiation only, chemotherapy-based regimen, and other)
Remains significant at p=0.05 level following FDR adjustment
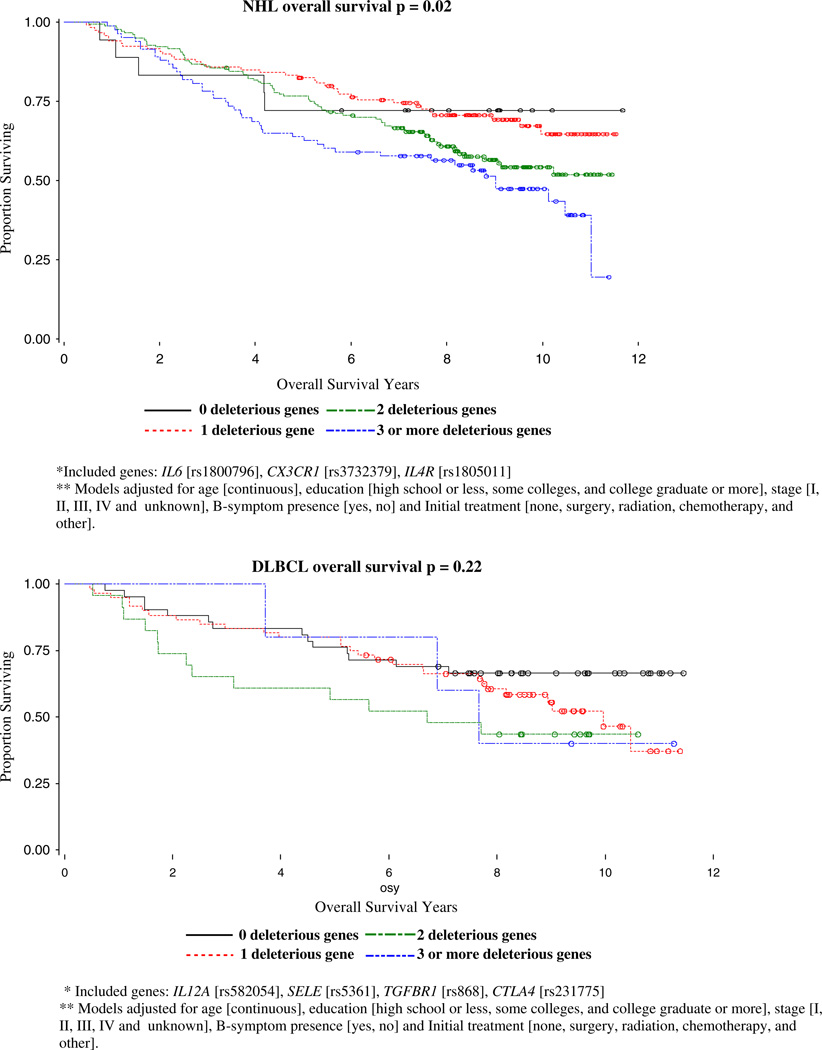
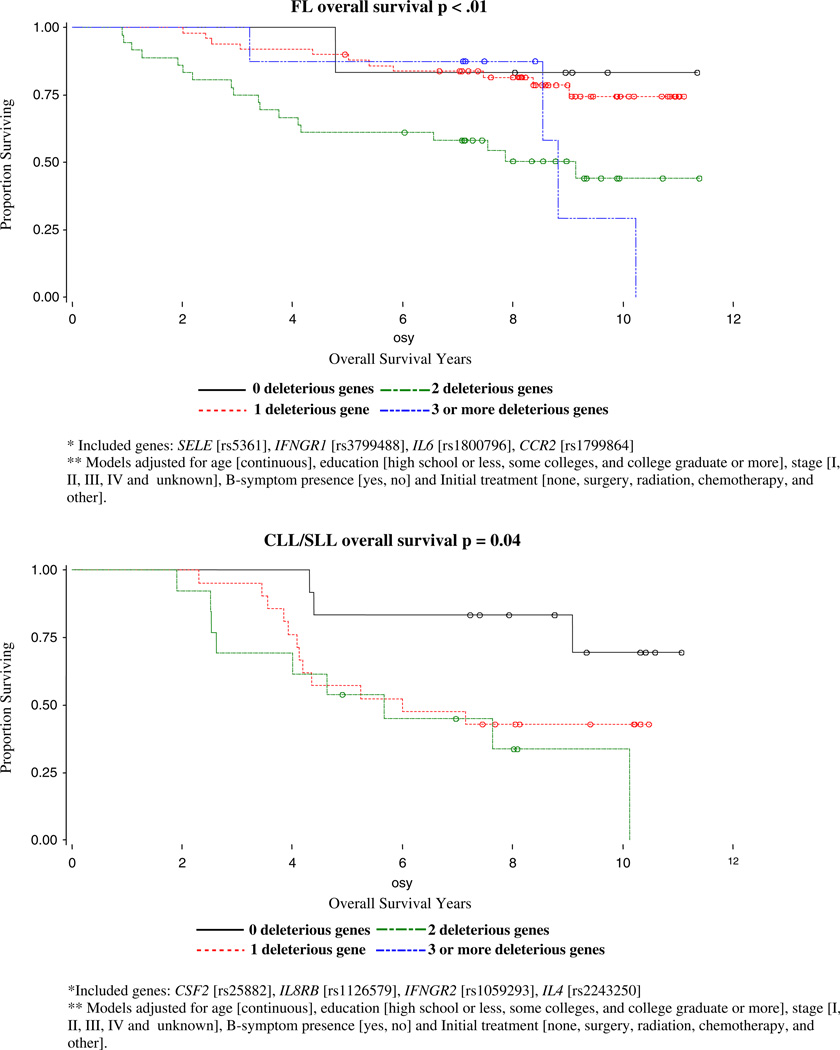
With backward selection for OS models, three out of 10 SNPs were identified as risk SNPs for NHL OS: IL6 (rs1800796), CX3CR1 (rs3732379), IL4R (rs1805011); the four out of 10 SNPs were identified as risk SNPs for DLBCL OS: IL12A (rs582054), SELE (rs5361), TGFBR1 (rs868), CTLA4 (rs231775); four out of 11 risk SNP for FL OS were identified as SELE (rs5361), IFNGR1 (rs3799488), IL6 (rs1800796), CCR2 (rs1799864); and four out of 8 SNPs were identified as risk SNPs for CLL/SLL OS: CSF2 (rs25882), IL8RB (rs1126579), IFNGR2 (rs1059293), IL4 (rs2243250). The homogeneity of OS curves by number of deleterious genotypes and p-values for log-rank tests for NHL overall and subtype cases are shown in Fig. 1. We identified significantly different effects of increasing deleterious genotypes for NHL overall (p=0.0155), for the follicular (p=0.0052) and CLL/SLL (p=0.0428) subtypes, but not for DLBCL (p=0.2175).
Fig. 1.
Kaplan-Meier overall survival curves by the number of deleterious genotypes for NHL cases, Connecticut, 1995–2001
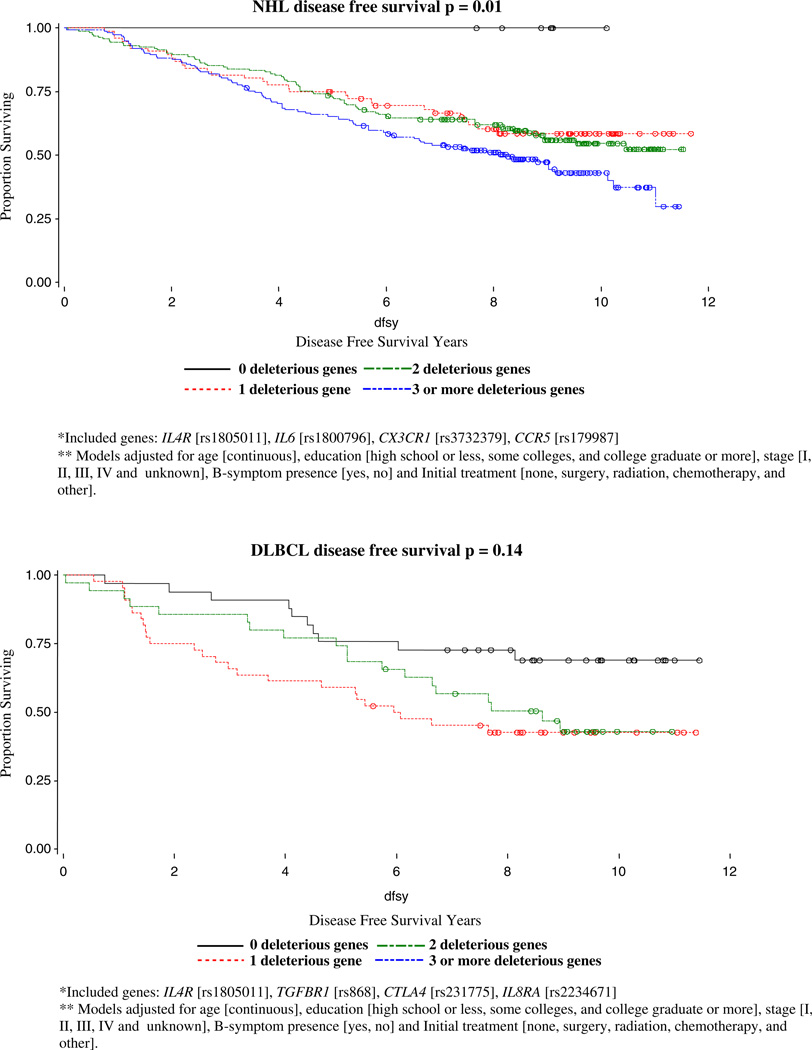
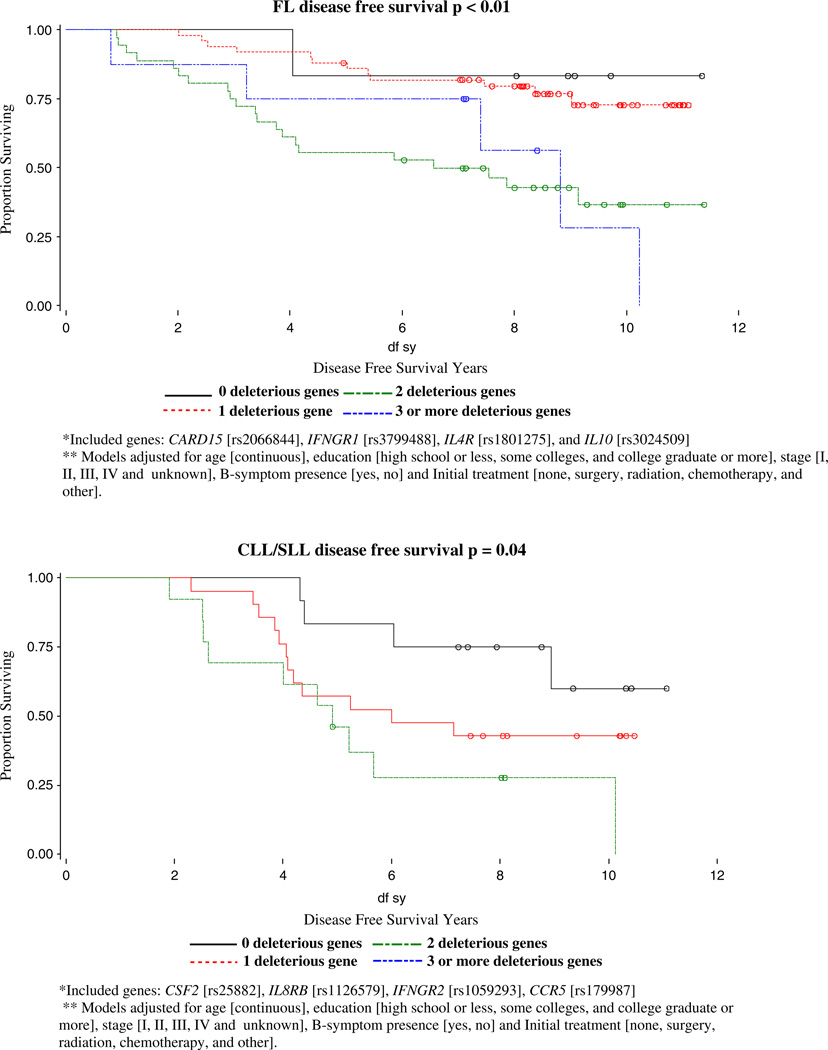
With backward selection for DFS models, four out of 8 SNPs were identified as risk SNPs for NHL DFS: IL4R (rs1805011), IL6 (rs1800796), CX3CR1 (rs3732379), CCR5 (rs179907); IL4R (rs1805011), TGFBR1 (rs868), CTLA4 (rs231775), IL8RA (rs2234671) were identified as the four risk SNPs for DLBCL DFS; CARD15 (rs2066844), IFNGR1 (rs3799488), IL4R (rs1801275), and IL10 (rs3024509) were identified as risk SNPs for FL DFS; and CSF2 (rs25882), IL8RB (rs1126579), IFNGR2 (rs1059293), CCR5 were identified as the four risk SNPs for CLL/SLL DFS. The homogeneity of DFS curves by number of deleterious genotypes and p-values for log-rank tests for NHL overall and subtype cases are shown in Fig. 2. We identified significantly different effects of increasing deleterious genotypes for NHL overall (p=0.0016), for follicular (p=0.0012), and CLL/SLL (p=0.0390) subtypes, but not for DBLCL (p=0.1373).
Fig. 2.
Kaplan-Meier disease-free survival curves by the number of deleterious genotypes for NHL cases, Connecticut, 1995–2001
Discussion
Using a population-based sample of NHL cases diagnosed from 1996 to 2000 and followed through 2008, we identified 11 SNPs from 9 cytokine and related immune regulation genes that were associated with overall survival for NHL and NHL subtypes, and 10 SNPs from 9 cytokine and related immune regulation genes that were associated with overall survival, relapse, or secondary cancer for NHL and NHL subtypes. From these SNPs, we then identified the strongest predictors of survival and combined them into a common carrier model that summed the number of deleterious genotypes. Combining the SNPs with clinical and demographic factors significantly increased the predictive ability of the model over a model restricted to clinical and demographic factors. We were able to identify models that were significantly different based on the number of deleterious genotypes for NHL and the investigated NHL subtypes.
For NHL overall, the clearest signal for both overall survival as well as disease free survival was due to a polymorphism in IL6. IL6 is a pleiotropic pro- and anti-inflammatory cytokine and tumor growth factor implicated in the pathogenesis of AIDS-related non-Hodgkin’s lymphoma and Kaposi’s sarcoma [KS]. This SNP has been shown to affect both the transcription and secretion of IL6 [14, 15]. Several studies have suggested the involvement of a disordered production of IL6 in lymphomas [15– 19] and elevated levels of IL6 have been observed in serum from patients with newly diagnosed lymphomas [20]. Patients with elevated serum levels of IL6 has also been shown to have a poorer overall survival rate as compared with patients with normal levels [21]. The significance of IL6 as a prognostic marker in our study is consistent with such reports. In addition to IL6, we also found a signal from CCR5 for NHL survival. The CCR5 gene encodes a chemokine receptor protein that plays an important role in many immune-related processes [14] and the variant alleles have been shown to increase the risk of developing AIDs-related NHL though the role in NHL survival has not been evaluated previously.
For the follicular subtype, we found that SNPs in IFNGR1, IL4R, IL8, MIF, and SELE were significant prognostic factors. In a similar analysis by Cerhan et al [22] of immune function genes and follicular lymphoma survival, they found that SNP markers from IL8, IL2, IL12B, and IL1RN were the most robust predictors of survival individually. We also found that IL8 is a strong predictor of overall survival as well as relapse and secondary cancers for follicular lymphoma cases. Interleukin-8 is produced by a wide variety of cells at inflammatory sites and acts on neutrophils stimulating degranulation and chemotaxis. The identified polymorphism [rs4073] has been associated with higher production of IL8, and IL8 production has been linked with tumor vascularization, metastatic phenotype, and poor prognosis [23–25] including progression in patients with early-stage chronic lymphocytic leukemia [26]. Our finding that follicular lymphoma patients with the homozygous variant genotype [and therefore presumed lower IL8 production] had a poorer survival, is consistent with the Cerhan et al [22] finding and suggests that higher IL8 levels and concomitant greater inflammation may play a protective role in follicular lymphoma. Our finding that IFNGR1, IL4R, SELE, and MIF may be other strong predictors of survival for follicular lymphoma in our study population should be replicated in future work.
We found that for DLBCL, a SNP in SELE [responsible for the accumulation of blood leukocytes at sites of inflammation] was significant for OS whereas SNPs in FCGR2A [involved in the process of phagocytosis and clearing of immune complexes] and TGFBR1 [transduces the TGF-beta signal from the cell surface to the cytoplasm] were significant for disease free survival. We also found that polymorphisms in IL8RB [mediates neutrophil migration to sites of inflammation], CCR5, and TGFBR1 were significant prognostic factors for CLL/SLL survival.
We previously published the association of most of these SNPs with risk of developing NHL from our population-based case-control study [7]. In evaluating the risk of NHL development, we found that SNPs in each of 3 Th2-related cytokines, IL4, IL5, and IL10, were significantly associated with an increased risk for NHL overall which was consistent with results from a pooled analysis. Subsequent analyses by NHL subtype showed that variant SNPs in IL10 were significantly associated with increased risk for DLBCL and follicular lymphoma. In that analysis, SNPs in IL6 and IL8 were not associated with risk of all NHL or follicular lymphoma, respectively. These findings suggest that although genetic variability in immune genes seems to be associated with lymphoma-genesis in general, the specific SNPs and genes involved in NHL etiology may differ from genes involved in prognosis, although more data will be needed to fully evaluate this hypothesis.
The findings from the combined deleterious models for NHL overall and NHL subtypes indicated that improvements could be made to prognostic models by including information on deleterious genotypes. However, when we included the same SNPs identified by another group in our models [22], we did not get significant results. Specifically, when we ran the model for follicular lymphoma including IL1RN, IL12RB, IL2, and IL8 in our prognostic model for follicular lymphoma survival, we achieved a p-value of 0.85 for the test of homogeneity for the likelihood ratio. This may be due to the heterogeneity of patients in the population study. The biological mechanism is a complex system and different subtypes of cancer may have different genetic markers. Another potential explanation for this difference is that we had more detailed data on prognostic clinical factors. Our inability to replicate previous results suggests that although we have significantly improved the predictors of survival in this study population, our results need to be replicated in larger study populations such as pooled cohorts. If our models are found to significantly predict survival in other study populations, this could indicate that the inclusion of genotyping data in the clinical assessment of NHL and NHL subtypes may provide some benefit to clinicians and patients.
An important strength of this study was the population-based ascertainment of newly diagnosed cases. This study is the largest and most comprehensive study of immune candidate SNPs in relation to NHL survival conducted to date. The value of the study also lies in the fact that the choice of SNPs was based on either functional data or prior associations with cancer or other immune-related diseases. Our statistical analyses were comprehensive, and we have been cautious to evaluate the robustness of our results to both false positives and false negatives. We have not, however, reached median survival for the cohort, and further follow-up could lead to other SNP associations. The treatment information for chemotherapy collected by CTR is not detailed and comprehensive. Moreover the information on relapse and secondary cancer might be incomplete especially among patients who were no longer Connecticut residents, which could cause our measure of DFS to be longer than an accurate measure of time to outcome. However, the information bias is unlikely to be associated with their genetic background, thus our observed associations on DFS may be biased towards the null due to this non-differential misclassification. In the DFS analysis, we also considered of deaths in addition to relapses and occurrences of secondary cancer as events instead of censorings, potentially making the OS and DFS analyses more similar than if deaths were censored. Finally, there could be confounding by comorbid conditions that are associated with these cytokine genotypes and that impact survival.
In order to investigate the generalizability of the study results, we compared our overall survival curve with that of 13,899 female NHL patients aged 21–84 diagnosed during 1996–2000 at 17 Surveillance, Epidemiology and End Results (SEER) registries [27]. The two survival curves were parallel with around 15% difference in survival rates throughout the follow-up period except during the first half year (median time between diagnosis and interview 4.5 months, and mean 7 months), during which 15.8% of SEER patients died, while none of the patients in our study died. Considering that 167 out of 1,122 [14.9%] identified cases were not able to be enrolled in the CT study because they died before interview [8], the overall survival of our case series is comparable to the survival observed by SEER. Our results might not apply to the most aggressive NHL cases with short-term survivals since there were limited cases in our study who died within a half year.
Our results support the hypothesis that inherited genetic variation in cytokine and related regulatory immune genes could influence survival in NHL, as well as the DLBCL, follicular, and CLL/SLL subtypes, and provide further evidence for the contribution of host factors, including the tumor microenvironment, in the prognosis of NHL and NHL subtype survival [28]. Our findings are also consistent with the emerging significance of the complex host genetic background in progression of cancer in general [29]. In the future, the clinical evaluation of host genetics in cancer patients could become a useful tool in tailoring individual patient management and therapy, and complement gene expression profiling as a prognostic tool [29]. Ultimately, the simultaneous evaluation of host and tumor factors would be expected to increase prognostic ability.
In summary, host genetic variability in immune genes, individually and in particularly in combination, appear to be associated with overall survival and disease free survival in NHL and NHL subtypes after accounting for clinical and demographic factors. In this regard, host immunogenetics represents a promising class of prognostic factors that warrant further evaluation. Furthermore, our analysis suggests that such prognostic factors may differ by subtype and should be evaluated accordingly.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH/National Cancer Institute [NCI], grant CA62006 from the NCI, by Hull Argall & Anna Grant 22067A from the Yale Cancer Center, and by Fogarty training grants 1D43TW008323-01 and 1D43TW007864-01 from the National Institute of Health [NIH]. This publication was made possible by CTSA Grant number UL1 RR024139 from the National Center for Research Resources [NCRR], a component of the NIH and NHL roadmap for medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR. This research was approved by the DPH HIC. Certain data used in this study were obtained from the Connecticut Department of Public Health. The authors assume full responsibility for analyses and interpretation of these data.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11764-010-0164-4) contains supplementary material, which is available to authorized users.
Contributor Information
Briseis Aschebrook-Kilfoy, School of Public Health, Yale University, New Haven, CT, USA; Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Department of Health and Human Services, Rockville, MD, USA.
Tongzhang Zheng, School of Public Health, Yale University, New Haven, CT, USA.
Francine Foss, Department of Medical Oncology, Yale University School of Medicine, New Haven, CT, USA.
Shuangge Ma, School of Public Health, Yale University, New Haven, CT, USA.
Xuesong Han, School of Public Health, Yale University, New Haven, CT, USA.
Qing Lan, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Department of Health and Human Services, Rockville, MD, USA.
Theodore Holford, School of Public Health, Yale University, New Haven, CT, USA.
Yingtai Chen, Cancer Institute/Hospital, Chinese Academy of Medical Siences, Beijing, China.
Brian Leaderer, School of Public Health, Yale University, New Haven, CT, USA.
Nathaniel Rothman, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Department of Health and Human Services, Rockville, MD, USA.
Yawei Zhang, School of Public Health, Yale University, New Haven, CT, USA.
References
- 1.Clarke C, O’Malley C. Chapter 28 Non-Hodgkin lymphoma. In: Ries LAG, Yong JL, Keel GE, Eisner MP, Lin YD, Horner M-JD, editors. Cancer survival among adults: U.S. SEER Program, 1988–2001, patient and tumor characteristics. Bethsda: National Cancer Institutes, SEER Program, NIH; 2007. pp. 235–242. [Google Scholar]
- 2.Han X, Kilfoy B, Zheng T, Holford TR, Zhu C, Zhu Y, et al. Lymphoma survival patterns by WHO subtype in the United States, 1973–2003. Cancer Causes Control. 2008;19(8):841–858. doi: 10.1007/s10552-008-9147-4. Epub 2008 Mar 26. [DOI] [PubMed] [Google Scholar]
- 3.Groves FD, Linet MS, Travis LB, Devesa SS. Cancer surveillance series: non-Hodgkin’s lymphoma incidence by histologic subtype in the United States from 1978 through 1995. J Natl Cancer Inst. 2000;92(15):1240–1251. doi: 10.1093/jnci/92.15.1240. [DOI] [PubMed] [Google Scholar]
- 4.The International Non-Hodgkin’s Lymphoma Prognostic Factors Project. A predictive model for aggressive non-Hodgkin’s lymphoma. N Engl J Med. 1993;329(14):987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 5.Balkwill F, Mantovani A. Inflammation and cancer: Back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 6.Clarke CA, Glaser SL. Changing incidence of non-Hodgkin lymphomas in the United States. Cancer. 2002;94(7):2015–2023. doi: 10.1002/cncr.10403. [DOI] [PubMed] [Google Scholar]
- 7.Lan Q, Zheng T, Rothman N, et al. Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood. 2006;107:4101–4108. doi: 10.1182/blood-2005-10-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morton LM, Holford TR, Leaderer B, Boyle P, Zahm SH, Zhang Y, et al. Cigarette smoking and risk of non-Hodgkin lymphoma subtypes among women. Br J Cancer. 2003;89(11):2087–2092. doi: 10.1038/sj.bjc.6601388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Holford TR, Leaderer B, Zahm SH, Boyle P, Morton LM, et al. Prior medical conditions and medication use and risk of non-Hodgkin lymphoma in Connecticut United States women. Cancer Causes Control. 2004;15(4):419–428. doi: 10.1023/B:CACO.0000027506.55846.5d. [DOI] [PubMed] [Google Scholar]
- 10.A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin’s lymphoma. The Non-Hodgkin’s Lymphoma Classification Project. Blood. 1997;89:3909–3918. [PubMed] [Google Scholar]
- 11.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, et al. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J Clin Oncol. 1999;17:3835–3849. doi: 10.1200/JCO.1999.17.12.3835. [DOI] [PubMed] [Google Scholar]
- 12.Packer BR, Yeager M, Burdett L, Welch R, Beerman M, Qi L, et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–D621. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scala G, Quinto I, Ruocco MR, Arcucci A, Mallardo M, Caretto P, et al. Expression of an exogenous interleukin-6 gene in human Epstein Barr virus B-cells confers growth advantage and tumorigenicity. J Exp Med. 1990;172:61–68. doi: 10.1084/jem.172.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smallwood L, Allcock R, van Bockxmeer F, Warrington N, Palmer LJ, Iacopetta B, et al. Polymorphisms of the interleukin-6 gene promoter and abdominal aortic aneurysm. Eur J Vasc Endovasc Surg. 2008;35(1):31–36. doi: 10.1016/j.ejvs.2007.08.021. Epub 2007 Nov 8. [DOI] [PubMed] [Google Scholar]
- 16.Yee C-Y, Biondi A, Wang X-H, Iscove NN, de Sousa J, Aarden LA, et al. A possible autocrine role for interleukin-6 in two lymphoma cell lines. Blood. 1989;74:798–804. [PubMed] [Google Scholar]
- 17.Takeshita M, Sumiyoshi Y, Masuda Y, Ohshima K, Yoshida T, Kikuchi M, et al. Cytokine[interleukin- la, interleukin- Ip, tumor necrosis factor a, and interleukin-6]-possessing cells in lymph nodes of malignant lymphoma. Pathol Res Pract. 1993;189:18–25. doi: 10.1016/S0344-0338(11)80112-3. [DOI] [PubMed] [Google Scholar]
- 18.Freeman GJ, Freedman AS, Rabinowe SN, Segil JM, Horowitz J, Rosen K, et al. Interleukin 6 gene expression in normal and neoplastic B-cells. J Clin Invest. 1989;83:1512–1518. doi: 10.1172/JCI114046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emilie D, Coumbaras J, Raphael M, Devergne O, Delecluse HJ, Gisselbrecht C, et al. Interleukin-6 production in high-grade B-lymphomas: correlation with the presence of malignant imniunoblasts in acquired immunodeficiency syndrome and in human immunodeficiency virus-seronegative patients. Blood. 1992;80:498–504. [PubMed] [Google Scholar]
- 20.Bataille R, Jourdan M, Zhang X-G, Klein B. Serum levels of interleukin 6. a potent myeloma cell growth factor, as a reflection of disease severity in plasma cell dyscrasias. J Clin Invest. 1989;84 doi: 10.1172/JCI114392. 008–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chopra GS, Chitalkar PG, Jaiprakash MP. Cytokines: as useful prognostic marker in lymphoma cases. MJAFI. 2004;60:45e49. doi: 10.1016/S0377-1237(04)80158-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerhan JR, Wang S, Maurer MJ, Ansell SM, Geyer SM, Cozen W, et al. Prognostic significance of host immune gene polymorphisms in follicular lymphoma survival. Blood. 2007;109(12):5439–5446. doi: 10.1182/blood-2006-11-058040. Epub 2007 Feb 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueda T, Shimada E, Urakawa T. Serum levels of cytokines in patients with colorectal cancer: possible involvement of interleukin-6 and interleukin-8 in hematogenous metastasis. J Gastroenterol. 1994;29:423–429. doi: 10.1007/BF02361238. [DOI] [PubMed] [Google Scholar]
- 24.Kitadai Y, Haruma K, Sumii K, et al. Expression of interleukin-8 correlates with vascularity in human gastric carcinomas. Am J Pathol. 1998;152:93–100. [PMC free article] [PubMed] [Google Scholar]
- 25.Nurnberg W, Tobias D, Otto F, Henz BM, Schadendorf D. Expression of interleukin-8 detected by in situ hybridization correlates with worse prognosis in primary cutaneous melanoma. J Pathol. 1999;189:546–551. doi: 10.1002/(SICI)1096-9896(199912)189:4<546::AID-PATH487>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 26.Molica S, Vitelli G, Levato D, Levato L, Dattilo A, Gandolfo GM. Clinicobiological implications of increased serum levels of interleukin-8 in B-cell chronic lymphocytic leukemia. Haematologica. 1999;84:208–211. [PubMed] [Google Scholar]
- 27.SEER Limited-Use 1973–2004—ASCII Text Data: Surveillance, Epidemiology, and End Results [SEER] Program [www.seer.cancer.gov] Limited-Use Data [1973–2004], National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch, released April 2007, based on the November 2006 submission.
- 28.Kuppers R. Prognosis in follicular lymphoma—it’s in the microenvironment. N Engl J Med. 2004;351:2152–2153. doi: 10.1056/NEJMp048257. [DOI] [PubMed] [Google Scholar]
- 29.Hunter KW, Crawford NP. Germ line polymorphism in metastatic progression. Cancer Res. 2006;66:1251–1254. doi: 10.1158/0008-5472.CAN-05-3705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.