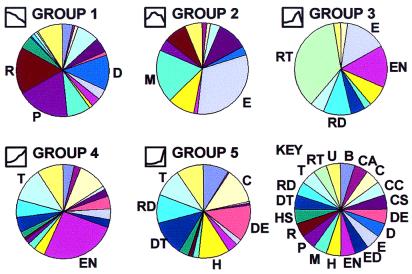
Figure 6.
Functional associations of gene clusters. Gene clusters varied remarkably in terms of major functional classifications of component genes. Key (Lower right) indicates major gene functional classifications: B, biosynthetic; CA, cell adhesion; C, catabolism-small molecules; CC, cell cycle; CS, cytoskeletal; DE, defense; D, DNA structure or replication; E, extracellular matrix; ED, endocytosis; EN, energy metabolism; H, homeostasis of the organism; M, morphogenetic; P, protein synthesis or processing; R, RNA synthesis or processing; HS, heat-shock proteins; DT, detoxification of exogenous substances; RD, protection against oxidative stress; T, transport; RT, retrotransposon; U, unknown function. The icons preceding the group names were derived from Fig. 4 and display the associated temporal expression profile. Group 1 expressed earlier in nephrogenesis was most notable for genes involved in DNA replication (D), RNA production (R), protein synthesis (P), and morphogenesis (M), consistent with an actively proliferating tissue. Group 2 (which peaked in midnephrogenesis) was most notable for genes of the extracellular matrix (E) as well as morphogenetic genes (M). Group 3 (with a peak in neonatal life) was dominated by retrotransposon transcripts (RT). Group 4 was most notable for transport (T) and energy metabolism (EN) related genes. Group 5 genes (significantly up-regulated in the adult vs. all previous times) was more heterogeneous and included genes specifying catabolic enzymes (C), defense and immune recognition (DE), homeostasis of the organism as a whole (H), detoxification (DT), oxidative stress (RD), and transport (T).