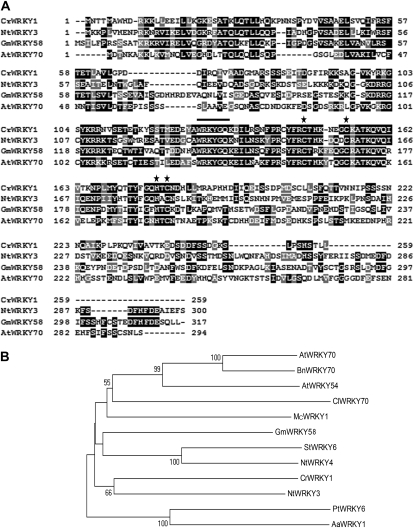
Figure 1.
Multiple sequence alignment and phylogenetic analysis of selected WRKY proteins. A, The deduced amino acid sequence of CrWRKY1 is aligned with homologs from N. tabacum (NtWRKY3; accession no. AF193770), G. max (GmWRKY54; accession no. EU375354), and Arabidopsis (AtWRKY70; accession no. AF421157). The WRKY signature motif (WRKYGQK) is indicated by the line, and the characteristic residues for the zinc-finger motif are marked by asterisks. B, A neighbor-joining phylogenetic tree of CrWRKY1 and selected WRKY domain proteins from other plant species constructed using the MEGA4 software. The statistical reliability of individual nodes of the newly constructed tree is assessed by bootstrap analyses with 1,000 replications. The WRKY proteins, their respective plant species, and GenBank accession numbers are as follows: AtWRKY70, Arabidopsis (AF421157; At3G56400); BnWRKY70, Brassica napus (FJ384113); AtWRKY54, Arabidopsis (NM_129637; At2G40750); ClWRKY70, Citrullus lanatus (GQ453670); McWRKY1, Matricaria chamomilla (AB035271); GmWRKY58, G. max (EU375354); StWRKY6, Solanum tuberosum (EU056918); NtWRKY3, N. tabacum (AF193770); CrWRKY1, C. roseus (HQ646368); NtWRKY4, N. tabacum (AF193771); PtWRKY6, Populus tomentosa × Populus bolleana (GQ377426); AaWRKY1, Artemisia annua (FJ390842).