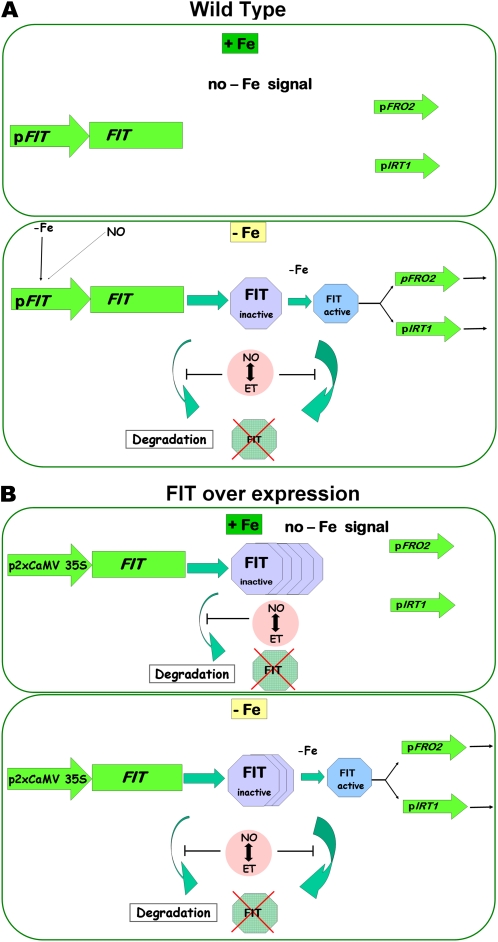
Figure 7.
Model summarizing the regulation of FIT activity in wild-type and HA-FIT Ox plants by multiple control steps. A, The wild type. In +Fe wild-type roots, FIT induction does not take place. It might be repressed by a negative regulator. Downstream targets of FIT, like FRO2 and IRT1, are not induced. In –Fe wild-type roots, FIT transcription is induced. The presumptive FIT repressor protein might be removed by –Fe. Subsequently, FIT protein is produced. Due to a –Fe signal, FIT is activated and promotes the induction of FRO2 and IRT1. The transition from inactive to active FIT could be achieved through FIT protein modification by the addition or removal of covalent modifications, or by a differential interaction with proteins present only at –Fe, such as bHLH038 and bHLH039 (Yuan et al., 2008), or perhaps both. FIT itself is degraded due to protein turnover (compare this work with Lingam et al., 2011; Sivitz et al., 2011). NO and ethylene (ET) increase the accumulation of FIT by counteraction of proteasomal FIT degradation (compare this work and Lingam et al., 2011). B, HA-FIT overexpression. Transcriptional control of HA-FIT is not relevant due to the 2xCaMV 35S promoter. In +Fe HA-FIT roots, HA-FIT is targeted by protein turnover. NO and ethylene increase the accumulation of FIT, probably by counteraction of proteasomal FIT degradation. In the absence of a –Fe activating signal, FRO2 and IRT1 targets are not induced. In –Fe HA-FIT roots, HA-FIT is activated by a –Fe signal and promotes the expression of FRO2 and IRT1. Further details in B are as in A. [See online article for color version of this figure.]