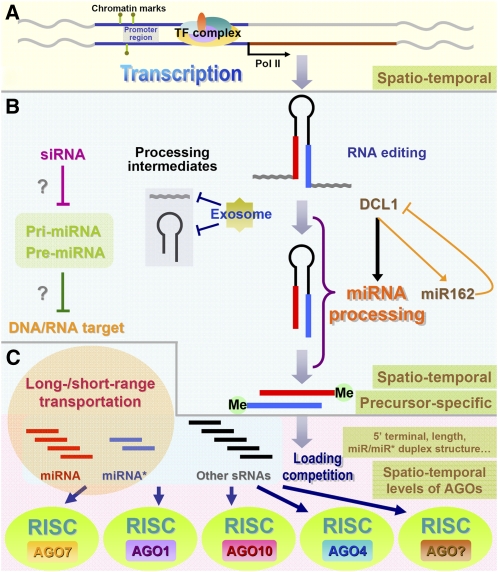
Figure 1.
Schematic presentation showing the dynamic nature of miRNA biogenesis in plants. A, RNA polymerase II (Pol II)-dependent transcription of the miRNA genes. Chromatin marks including DNA methylation and histone modifications, and the combinatory regulation of many TFs, together contribute the spatiotemporal expression patterns of the miRNA genes in plants. B, Processing of miRNA precursors and miRNA maturation. RNA editing on the miRNA precursors plays a role in changing the original sequence information encoded by the miRNA gene loci. The exosome was suggested to be implicated in digesting the processing intermediates from the miRNA precursors, ensuring relatively high processing efficiency. The processing efficiency also shows a high precursor sequence-specific dependence. Methylation at the 3′ ends of the miRNA/miRNA* duplex (“Me” here represents the methyl group) is crucial for the stabilization of miRNA and miRNA*. A feedback circuit between DCL1 and miR162 exists within the processing procedure. Moreover, the siRNAs complementary to specific pri-miRNAs and pre-miRNAs exhibit a potential repressive role in miRNA processing. More interestingly, the pri-miRNAs and pre-miRNAs may possess their own targets. C, Sorting into the AGO-associated miRISCs. A drastic loading competition may exist among miRNAs, miRNA*s, and other sRNA species. Not all the miRNAs are incorporated into the AGO1 complex. The 5′ terminal composition and the sequence length of the miRNAs, the structure of the miRNA/miRNA* duplex, and other undetermined factors have a significant influence on the loading patterns of the mature miRNAs.