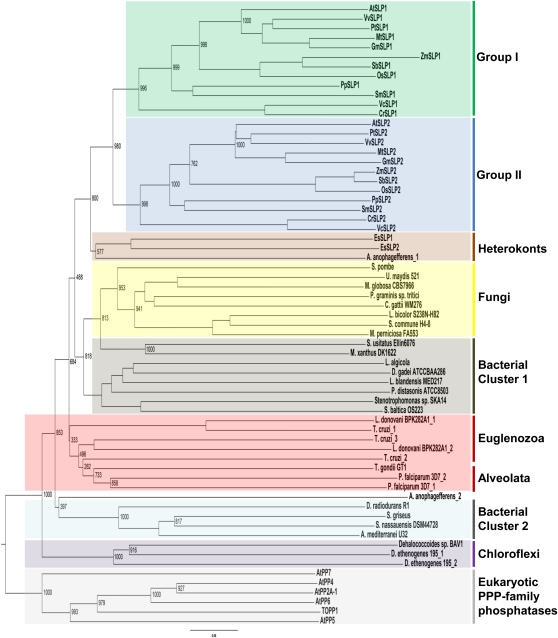
Figure 1.
Phylogenetic analysis of select SLP phosphatases from prokaryotic ancestors to conserved eukaryotic homologs. A rooted tree was generated through the comparison of homologous full-length SLP sequences with the exception of SmSLP2 (partial sequence), obtained by BLASTP analyses. Select eukaryotic Arabidopsis PPPs were used as an outgroup to anchor the tree. Homologous SLPs were aligned using ClustalX, and the bootstrap tree was generated using the neighbor-joining function of ClustalX. The group I and group II designations were assigned based on prediction algorithms that identified either a chloroplast or cytosol subcellular localization (Supplemental Fig. S1). Numbers represent bootstrap values for that node.