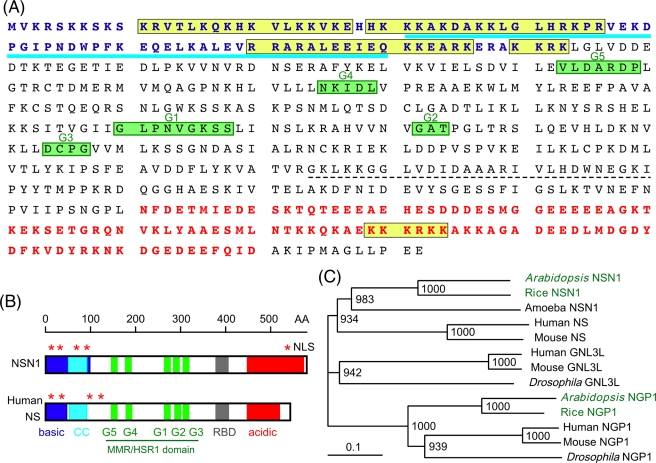
FIGURE 1:
Functional motifs and phylogenetic tree of Arabidopsis nsn1. (A) Amino acid sequence of NSN1 (At3g07050). Blue, basic domain (basic); blue underline, coiled-coil domain (CC); yellow boxes, nuclear localization signals (NLS); green boxes, GTP-binding motifs (G1–G5); dashed underline, RNA-binding domain (RBD); red, acidic domain (acidic). (B) Comparison of Arabidopsis NSN1 with human NS. The GTP-binding motifs (G1–G5) in NSN1 are arranged in a circularly permuted manner known as the MMR/HSR1 domain. Five NLS motifs are indicated by asterisks. Shaded areas correspond to the structural domains indicated in A. (C) Phylogenetic tree of NSN1 homologues. Human and mouse contains three NS-related genes (NS, GNL3L, and NGP1), whereas Arabidopsis and rice have two (NSN1 and NGP1). The amoeba Dictyostelium has only one NSN1 gene. Peptide sequences used for phylogenetic analysis include At3g07050 (AtNSN1), At1g52980 (AtNGP1), Os01g0375000 (rice NSN1), Os03g0352400 (rice NGP1), AAV74413 (human NS), AAH11720 (human GNL3L), Q13823 (human NGP1), AAO19472 (mouse NS), NP_932778 (mouse GNL3L), NP_663527 (mouse NGP1), Q8MT06 (Drosophila NGL3L), EAL25205 (Drosophila NGP1), and Q54KS4 (amoeba NSN1). Bootstrap values from 1000 trials are displayed on nodes.