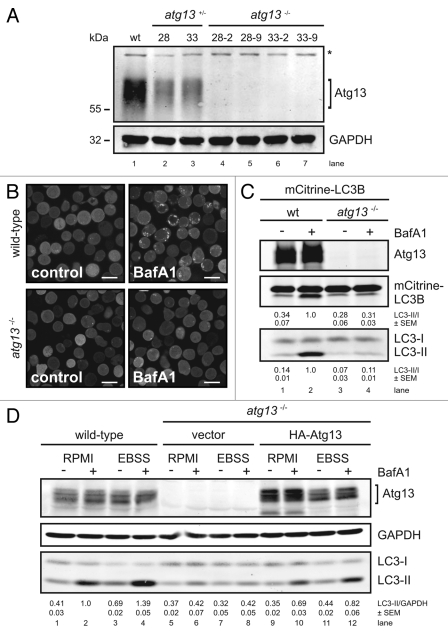
Figure 1.
Generation of Atg13-deficient DT40 cells. (A) Atg13-deficient DT40 B cell lines (atg13−/−) were generated by targeted disruption of both atg13 alleles. Successful targeting was confirmed by genomic PCR using primers specific for wild-type, hisD- or bsr-targeted alleles (see Fig. S1 for details). Equal amounts of protein from cleared cellular lysates of either wild-type (wt) cells (lane 1), atg13+/− clones (lanes 2–3) and atg13−/− clones (lanes 4–7) were analyzed for Atg13 and GAPDH by immunoblotting. The asterisk indicates an unspecific background band. (B) Wild-type and atg13−/− DT40 cells, retrovirally transfected with cDNA encoding mCitrine-LC3B, were treated with 10 nM bafilomycin A1 (BafA1) or DMSO (control) for 6 h and directly visualized by confocal laser scanning microscopy (bars: 10 µm). (C) Cleared cellular lysates of cells described in (B) were subjected to anti-Atg13 and anti-LC3 immunoblotting. LC3-II/LC3-I ratios are represented as mean values of three independent experiments ± SEM (D) atg13−/− cells reconstituted with HA-tagged full-length chicken Atg13 isoform A (lanes 9–12) were incubated in normal growth medium (RPMI) or starvation medium (EBSS) for 1 h in the presence or absence of 10 nM (BafA1). Equal protein amounts from cleared cellular lysates were analyzed for Atg13, GAPDH and LC3 by immunoblotting. As control, wild-type cells (lanes 1–4) and atg13−/− cells reconstituted with empty vector (lanes 5–8) were analyzed in parallel. LC3-II/GAPDH ratios are represented as mean values of three independent experiments ± SEM.