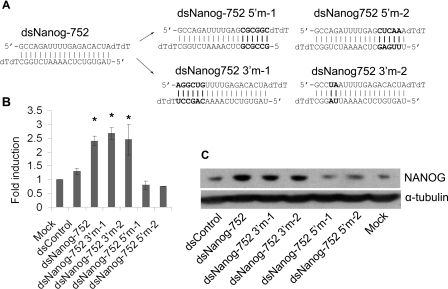
Figure 3. Sequence requirement for dsNanog-752.
(A) Mutations to the 5′-end (relative of the antisense strand) of dsNanog-752 resulted in duplexes dsNanog-752-5′m-1 and dsNanog-752-5′m-2. Mutations to the 3′-end of dsNanog-752 created mutant derivatives dsNanog-752-3′m-1 and dsNanog-752-3′m-2. The mutated bases are shown in bold. (B) NCCIT cells were transfected with 50 nM of the indicated saRNA molecules for 96 h. Expression of NANOG was accessed by quantitative RT–PCR and normalized to GAPDH. Results are fold changes relative to mock transfections (means±S.D. for two independent experiments). *P<0.05 compared with the mock. (C) NCCIT cells were transfected as in (B) and NANOG protein levels were detected by immunoblot analysis. α-Tubulin served as a loading control.