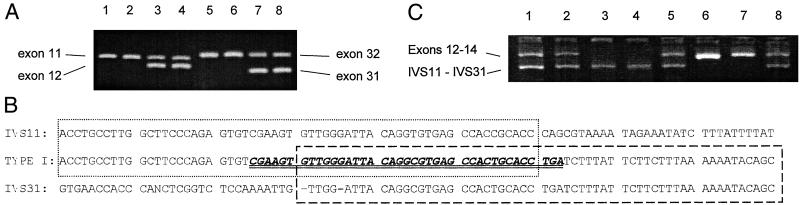
Figure 2.
(A) PCR analysis of the type I deletion. Lanes 1–4, biplex PCR of exons 11 and 12; lanes 5–8, biplex PCR of exons 31 and 32. The type I homozygotes in lanes 1, 2, 5, and 6 lack exons 12 and 31; lanes 3, 4, 7, and 8 are normal controls. (B) Sequence analysis of the type I deletion breakpoint. IVS11 and 31 show experimentally determined sequences from introns 11 and 31 in a 5′ to 3′ direction. The sequence determined from the type 1 breakpoint PCR product is shown between the intronic sequences. The dotted box shows homology between IVS11 sequence and the 5′ end of the type 1 sequence. The dashed box shows homology between the type 1 sequence and IVS31. The overlap between these two regions of homology is the likely recombination site. The double-underlined italic bold motif in the type 1 sequence was found to be highly homologous to Alu repeat sequences of various subfamilies. An unidentified nucleotide is designated N in the IVS31 sequence. (C) PCR analysis of the type I deletion. The larger product is a 1.3-kb fragment amplified from exons 12–14 of the wild-type FANCA gene, and the smaller product is the 715-bp fragment from the type I deletion. Lanes 1, 2, 5, and 8, type I heterozygotes; lanes 6 and 7, controls; lanes 3 and 4, type I homozygotes.