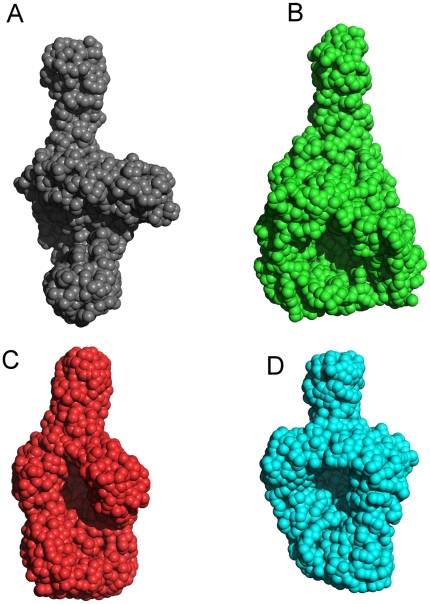
Figure 4. Ab initio models of ECAM generated by SAXS.
Each model results from averaging 10 individual models calculated by the program GASBOR using: (A) native ECAM, (B) methylamine-treated, (C) chymotrypsin-treated, and (D) elastase-treated ECAM. Note the appearance of a central cavity in all of the treated forms of the molecule. GASBOR was used in “user" mode, following default options, except for the total number of residues, which corresponded to total ECAM (1653 residues). The envelopes are based on the p(r) funtions shown on Fig. 3, and the GNOM files generated were used as input for GASBOR. The models are drawn to scale.