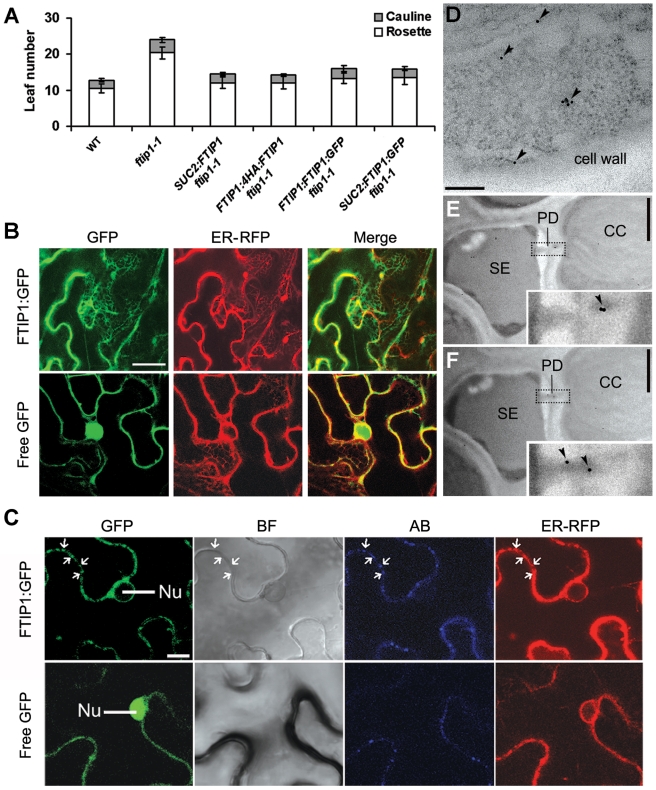
Figure 3. Subcellular localization of FTIP1.
(A) Flowering time of various transgenic plants grown under LDs. Error bars indicate SD. (B and C) Subcellular localization of FTIP1:GFP and free GFP in N. benthamiana leaf epidermal cells. (B) As compared to free GFP, FTIP1:GFP is mostly colocalized with an ER marker. (C) Both FTIP1:GFP and callose are enriched in the same regions at the cell wall (arrows). GFP, GFP fluorescence; ER-RFP, RFP fluorescence of an ER marker [17]; Merge, merge of GFP and RFP; BF, bright field image; AB, aniline blue staining; Nu, nucleus. Bars: (B), 20 µm; (C), 10 µm. (D–F) Analysis of 4HA:FTIP1 localization in CC-SE complexes in the first rosette leaves of 15-d-old FTIP1:4HA:FTIP1 ftip1-1 by immunogold electron microscopy. (D) 4HA:FTIP1 is localized in the phloem companion cell. Arrowheads indicate the locations of gold particles. (E,F) 4HA:FTIP1 is localized in the plasmodesma that connects a CC with a SE in two continuous sections. Arrowheads in insets show the location of gold particles in enlarged PD regions. SE, sieve element; CC, companion cell; PD, plasmodesma. Bars: (D), 250 nm; (E and F), 1 µm.