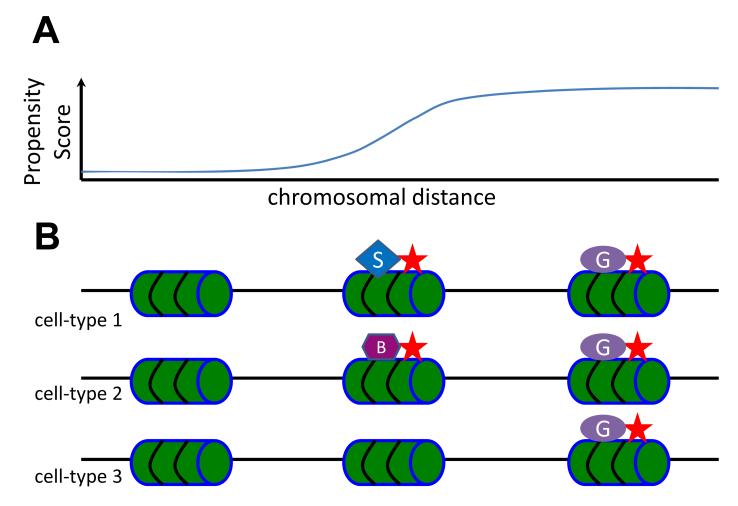
Figure 1.
A model for the role of the genomic sequence in guiding genome-wide epigenetic patterns. (A) The intrinsic association between an epigenetic mark and a genome locus is sequence-dependent and can be quantified by a propensity score. (B) An epigenetic mark (represented by the stars) is constitutively recruited to regions with high propensity scores, mediated by interaction with general factors (represented by the ovals) that target distinct sequence features. These features are usually degenerative and are the main determinants of the propensity scores. On the opposite end, this mark is excluded from regions with low propensity score. In the middle range, the epigenetic pattern is highly variable among different cell type. Occupancy can be enhanced or inhibited due to interactions with many cell type specific factors (represented by other shapes).