Figure 2.
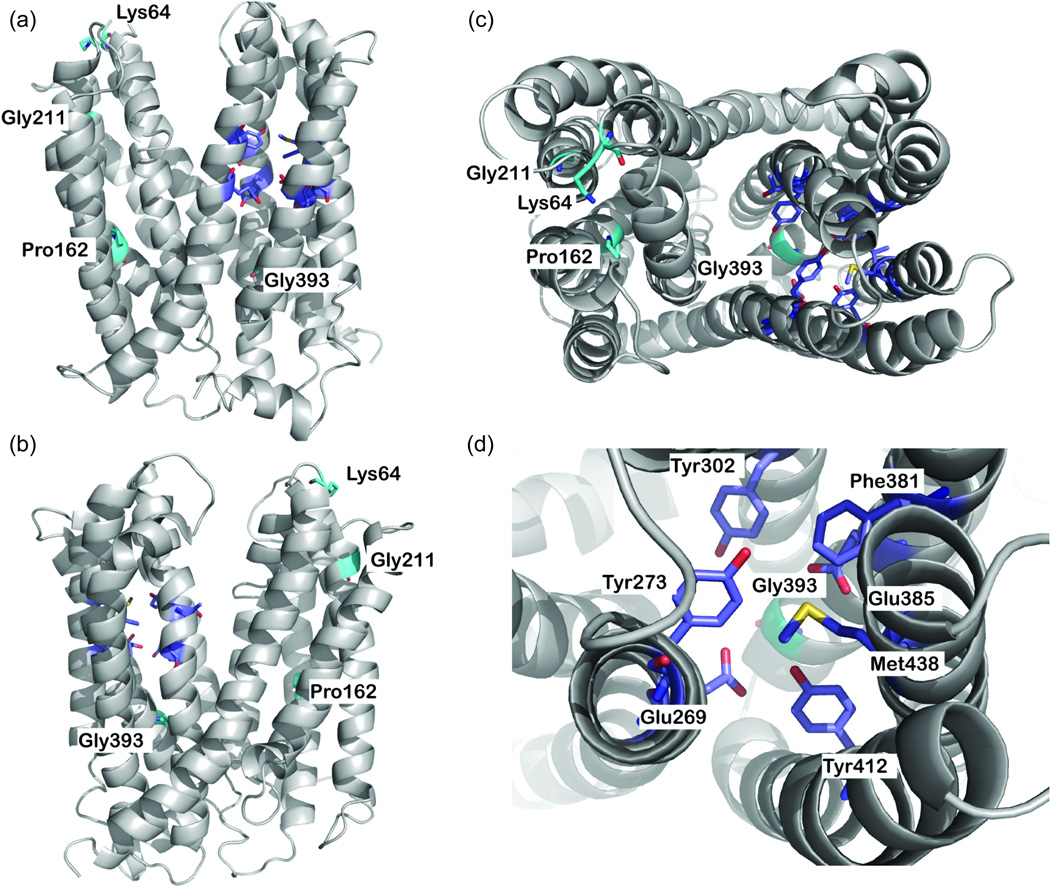
MATE2-K comparative model. (a,b) Side views and (c) top view of the predicted structure of MATE2-K. The backbone atoms of the model are depicted as a gray ribbon. Key residues are displayed as sticks; residues constituting the nonsynonymous variants are shown in cyan, and the putative proton binding site is shown in magenta. Oxygen, nitrogen, and hydrogen atoms are shown in red, blue, and white, respectively. (d) Top view of the putative proton binding site. In this model, two nonsynonymous variants, Pro162Leu (c.485C>T) and Gly393Arg (c.1177G>A), were discovered via Pharmacogenomics of Membrane Transporter (PMT) Projects (http://www.pharmacogenetics.ucsf.edu), and two nonsynonymous variants, Lys64Asn and Gly211Val, were previously reported by Japanese investigators.9 Also see Supplementary Figure S1 online for the putative secondary structure of MATE2-K showing the positions of four nonsynonymous variants and nine synonymous variants.
