Abstract
Plant lipid droplets are found in seeds and in post-embryonic tissues. Lipid droplets in seeds have been intensively studied, but those in post-embryonic tissues are less well characterised. Although known by a variety of names, here we will refer to all of them as lipid bodies (LBs). LBs are unique spherical organelles which bud off from the endoplasmic reticulum, and are composed of a single phospholipid (PL) layer enclosing a core of triacylglycerides. The PL monolayer is coated with oleosin, a structural protein that stabilizes the LB, restricts its size, and prevents fusion with adjacent LBs. Oleosin is uniquely present at LBs and is regarded as a LB marker. Although initially viewed as simple stores for energy and carbon, the emerging view is that LBs also function in cytoplasmic signalling, with the minor LB proteins caleosin and steroleosin in a prominent role. Apart from seeds, a variety of vegetative and floral structures contain LBs. Recently, it was found that numerous LBs emerge in the shoot apex of perennial plants during seasonal growth arrest and bud formation. They appear to function in dormancy release by reconstituting cell-cell signalling paths in the apex. As apices and orthodox seeds proceed through comparable cycles of dormancy and dehydration, the question arises to what degree LBs in apices share functions with those in seeds. We here review what is known about LBs, particularly in seeds, and speculate about possible unique functions of LBs in post-embryonic tissues in general and in apices in particular.
Keywords: 1,3-β-glucanase; bud dormancy; GH17 family protein; plant lipid body; plasmodesmata; seed dormancy; shoot apex
Lipid bodies
It has been known for over a century that lipid droplets are common constituents of plant cells. For example, as early as 1882, Sachs observed lipid droplets in parenchyma cells of axils in the root and the shoot while in 1888 they were noted in seeds by Walker.1 The literature refers to lipid droplets with a variety of names including fat droplets, lipid bodies, lipid globules, lipid spherules, lipid vesicles, lipoids, oil droplets, oleosomes, or sphersosomes. For simplicity we will refer to all of them as lipid bodies (LBs). As seed LBs are by far the best studied,2-4 they form an indispensible reference point for the investigation of plant LBs in general. LBs are minute membrane-bound organelles that range in size from about 0.5 to 2.5 µm,4-7 and which are pinched off from the endoplasmic reticulum (ER).8-10 Yatsu and Jacks11 provided evidence that in peanut seeds they consist of a core of neutral lipids bounded by a monolayer of phospholipids (PLs). Over time the picture emerged that virtually all plant LBs are composed of a core of non-polar lipid triacylglyceride (TAG) and a monolayer of amphipathic lipids.4,6,14 This configuration is relatively stable as the acyl moieties of the PLs interact with the TAG core while their hydrophilic head groups face the cytosol. TAGs and PLs are both synthesized by enzymes in the ER,2 and the enzyme that facilitates the last step in TAG synthesis, diacylglycerol acyl-transferase (DAG AT), is exclusive to the ER.12 Following synthesis, TAG is deposited between the two PL layers of the ER membrane, and diffuses to a specialized budding-site where LBs emerge for release into the cytosol.12-14 The smallest LBs may fuse until they reach their characteristic final size.15 During seed germination LBs become depleted when TAGs are metabolically processed to provide energy and carbon skeletons for growth. This involves breakdown of TAG to fatty acids, β-oxidation and subsequent processing in the glyoxylate/citric acid cycles of LB-associated glyoxisomes, conversion of succinate to malate in glyoxysome-associated mitochondria, and eventually hexose production by gluconeogenesis in the cytoplasm.5,16-18 Not surprisingly, LBs were traditionally regarded as organelle the exclusive function of which was to store oil reserves for germination.2,4-6,19 However, this picture is currently changing as LBs also appear to be involved in signaling processes, as will be discussed below.
Oleosin: LB stabilizer and marker
Despite the relative stability provided by the presence of a PL monolayer between the TAG core and the cytosol, it remains a most remarkable feature of LBs that they resist fusion and retain their small size. Early on it was proposed that the maintenance of LB integrity requires the presence of proteins that physically support the PL monolayer.11 Subsequent investigations confirmed this conjecture and revealed that all plant LBs are richly decorated with an LB-specific protein, named oleosin after the oleosome on which it was first identified.20 As a result, oleosin became regarded as a true LB marker.2-4,6,19,21,22 Oleosin, quantitatively spoken by far the most prominent LB protein, comes in two immunologically distinct weight-forms (L and H) which coexist in all LBs.5,23 The H-form differs from the L-form by having an extra stretch of amino acid residues at the C-terminal domain.2,5,23 Most oleosins range in size from 15 to 26 kDa,5,23,24 but sizes of up to 45 kDa have been reported. 25,26. In Arabidopsis, 16–18 oleosin genes have been identified, of which only five are exclusively expressed in seeds.12,15,25 The expression of oleosin genes is regulated at the transcriptional level and is under spatial and developmental regulation.27,28
Oleosins are highly unusual proteins. First, they do not have enzymatic domains and are rather inert structural proteins whose major function is to stabilize the LB. All oleosins have a thumbtack-like architecture,17 with three structural domains that are vital to its function in LB-stabilization: a variable N-terminal domain and a C-terminal α-helix that flank an exceptionally long central hydrophobic stretch of 70–80 amino acid residues.2,24-26,29 The central hydrophobic stretch forms a hairpin that stably anchors the protein in the TAG core of the LB, while the flanking domains are spread out on the surface of the phospholipid monolayer.30-32 The hairpin-bend occurs at the position of a Proline knot (Pro knot), a conserved motif that is required for LB targeting.2,33 Individual oleosins can form oligomeric associations13,14 and at the LB they interlock to give rise to a stable oleosin cage that shields off the PL monolayer from those of adjacent LBs and cytosolic constituents.21,34 By steric hindrance and electrostatic repulsion through its strongly negative charges, the oleosin cage prevents individual LBs from coalescing and fusing, a danger that is particularly imminent during the compaction process that accompanies cellular dehydration in maturing orthodox seeds.2,4,6,14,15,35 That oleosins can prevent fusion was experimentally demonstrated in experiments with artificial liposomes.36
The biological function of the cage was further demonstrated in transgenic plants where oleosin levels were strongly diminished. Ablation or attenuation of the major seed oleosin (OLE-1) in Arabidopsis by RNAi or T-DNA insertion resulted in malformed embryos with large LBs and a delayed germination.15 Similarly, RNAi knockdown of oleosins in soybean yielded fused LBs, disrupted cellular organization and impeded germination.37 Collectively, this provided further evidence that oleosins act as a surfactant37 which prevents LB fusion, and showed that this property is crucial for a timely germination. The advantage of small LBs is presumably that a high surface/volume ratio optimizes binding of cytoplasmic lipases during lipolysis, possibly at the C-terminal α–helix,2 thereby facilitating the rapid release of energy during post-germinative growth.19,38-40 It is telling that in cases where such rapid-release function is unnecessary, the LBs are not restricted in size. For example, the fatty mesocarp of fruits like avocado, olive and palm store oils to attract animals for seed dispersion and have large (10–20 µm) fused lipid droplets that lack oleosins.33,41 During seed germination LBs become emptied, resulting in deflated LBs. Subsequent collapse may result in the appearance of myelin-like structures formed by apposition of monolayers.11 LBs in various stages of deflation may fuse with the vacuole2,42 while H-oleosins and caleosin, a minor LB protein, become ubiquitinated for degradation.43
Oleosins are synthesized by polyribosomes3 at specialized ER domains.44 Although a specific signal sequence is lacking31,45 they target the ER via a signal recognition particle (SRP). The central hydrophobic stretch then functions as an aspecific sequence that is recognized by a hydrophobic pocket in the SRP.12,46 Subsequently, the oleosin is directed to a specialized microdomain at a distal tip of the tubular ER, where LBs form.2,10 Upon entry into the budding LB they may undergo a change in folding.13 Even though the Pro knot motif is not necessary for insertion into the ER, it is indispensible for the stable anchoring of the protein in the TAG core of the LB.31 In addition, it suffices to target oleosin to an artificial oil emulsion.7,36 Once regarded as a true marker for seed storage LBs, oleosin has been found in non-storage tissue like floral tapetum,2 pollen25,47 as well as a wide variety of vegetative plant tissues.14 Moreover, oleosins are not to be restricted to seed plants, as the moss Physcomitrella possesses oleosin genes and oleosin-coated LBs.48
Caleosin: LB stabilizer and signaling protein
In addition to oleosin, some minor proteins have been identified in LB fractions.5,49,50 Their discovery nuanced the original concept of the plant LB as an inert cellular organelle that exclusively served TAG storage. One of these minor proteins is caleosin,19 called Sop1 in sesame49 and ATS1 in Arabidopsis.7 Caleosin has three structural domains similar to those of oleosin, including a Pro knot motif, albeit less conserved.12 Association with the LB is mediated by the amphipathic α-helical stretch.44,49 Caleosin potentially contributes to LB stability, but its primary role might be in cytoplasmic signaling, as it possesses an N-terminal hydrophilic domain with a Ca2+-binding motif and a few phosphorylation sites at the hydrophilic C-terminal domain.49 A Ca2+-binding domain consists of a conserved EF-hand, which is a helix-loop-helix structural domain. Such EF-hands are typically found in the members of a large family of calcium-binding proteins.51 Ca2+-binding at EF-hands results in a conformational change in the particular protein, which then exposes a binding site for a target molecule. Caleosin has a unique position among the Ca2+-binding proteins as it has only one EF-hand, while normally a pair of hands creates a hydrophobic cup that accommodates two calcium ions.19 It was speculated therefore that caleosins may form cis or trans dimeric associations with other caleosins, for example to promote fusion of LBs with vacuoles for subsequent degradation.19,42,44 In Arabidopsis, seven caleosin-like genes are identified, only one of which is strongly expressed in seeds. Once thought to be specific of seed LBs, caleosins are also found associated with LBs in post-embryonic tissues including root tips,44 pollen47,74 and shoots.52 In addition, and in contrast to oleosin, caleosin is not restricted to plants as it is also found in fungi.6 The peroxygenase activity of caleosin may suggest that it is associated with phytooxylipin biosynthesis and defense responses.52 Notably, RD20 (Responsive to Dehydration, 20), a LB-associated caleosin in Arabidopsis, which is absent in seeds and roots, functions in the stress-signaling pathway of the shoot.53
Steroleosin: enzyme and signaling protein
A second minor protein found in LB fractions is steroleosin, called Sop2 in sesame,50,54 which is a membrane-associated NADP+-binding sterol dehydrogenase that belongs to a super-family of pre-signal proteins.50,55 The Pro knot motif, which in oleosins and caleosins typically presides in the central hydrophobic domain, is displaced in steroleosin to a hydrophobic N-terminal domain.36 This N-terminal domain anchors steroleosin in the TAG core. Perhaps due to this, and unlike oleosins and caleosins, steroleosins cannot stabilize artificial LBs.36 Sterol-binding dehydrogenases are involved in signal transduction in various plant tissues, and in seeds it is proposed to specifically facilitate mobilization of LBs during germination.50 Like caleosins, steroleosins are not seed-specific and are also present in post-embryonic tissues.14 In Arabidopsis, eight steroleosine genes are found with variable sterol-binding domains, suggesting that steroleosins have diversified to bind different regulatory sterols during the initiation of distinct signaling events, perhaps in a tissue-specific way.50 For example, hydroxysteroid dehydrogenase 1 (HSD1) in Arabidopsis is proposed to play a role in the regulation development by acting as a mediator of brassinosteroids signaling.56 Like in case of oleosin and caleosin, a cleavable signal sequence and post-translational modification is not present in steroleosin.50
Peripherally associated proteins: enzymes and signaling proteins
The LB storage paradigm initially might have predisposed to the view that only the structural protein oleosin and its various isoforms are canonical LB proteins, as they are specifically associated with LBs. Oleosin might be the only exclusive LB marker, but nonetheless caleosin and steroleosin are characteristically associated with LBs. In addition, however, LB fractions might reveal the presence of small amounts of other lipid- or protein-associated polypeptides. Often, these are regarded as cytoplasmic contaminants, and rigorous isolation procedures have been developed to remove them. As pointed out by Tzen et al.,5 a consequence of such rigid procedures is that not only cytoplasmic contaminants are washed out but also proteins that are non-covalently attached to the LB periphery. As it cannot be excluded that such proteins serve important biological functions, their presence warrants further investigation. For example, peripherally associated proteins may participate in the biosynthesis or degradation of oil, and include ER-associated enzymes, glyoxisome receptors, lipase, putative lipase receptors5,57 as well as unanticipated enzymes and signaling molecules. An example of a LB-associated enzyme is RcOBL1 in the endosperm of castor bean.58 RcOBL1 has acid lipase activity, and shares homology with lipases in taxonomically diverse species including plants, fungi and mammals.58 RcOBL1 is relatively amphipathic, but it lacks a prominent hydrophobic region like the one present in oleosins. Nonetheless, it was immunologically localized at the surface of the LBs, showing that is was not a cytoplasmic contaminant, and it could hydrolyze a range of TAGs.58 Lipase is an important enzyme as it acts by catalyzing the first step in the breakdown of TAGs, resulting in the release of free fatty acids and glycerol. The free fatty acids are then processed for catabolism by β-oxidation and conversion to sugar (see above). For example, in Arabidopsis seeds five lipase homologs are present at LBs.59 LBs might also possess patatin-like proteins, as shown in cucumber where a patatin-like protein with phospholipase A2 activity60 is believed to function in lipid mobilization and signaling.61 Similarly, developing seeds of Arabidopsis express the gene SUGAR-DEPENDENT1 (SDP1), the protein of which is localized to the LB surface.59 SDP1 possesses a patatin-like acyl-hydrolase domain that in vitro hydrolyzes TAGs and DAGs.59 Interestingly, LBs from dry seeds are resistant to attack by lipases and phospholipase A2 digestion, which requires imbibition.62,63 Thus, there might be a considerable time-lag or waiting period between lipase and phospholipase A2 production and digestive function, which is terminated only when rehydration allows the enzymes to access their substrate.
The perennial shoot apex: signaling through plasmodesmata
The question if LBs are present in shoot apices has not received much attention so far. This might not come as a surprise considering that LBs were regarded as organelles that serve long-term storage of TAGs, which does not seem to be of particular importance in the continuously propagating meristematic cells of the apex. However, in case of temperate perennials there are clear parallels with orthodox seeds as both, apices and seeds, undergo a transition to dormancy, involving multiple structural and physiological changes, including the emergence of numerous LBs (Fig. 1A‑D).64-70 It has not been investigated if, and to what degree LBs in dormant apices of perennial plants resemble LBs in seeds. For example, the question if apices express oleosin genes to equip their LBs has not been experimentally addressed. We have some evidence that dormant apices of Betula (birch) and Populus (poplar), which contain small (0.5–2.0 µm) LBs, indeed possess oleosins.67 In the poplar genome (Populus trichocarpa) eight oleosin genes have been identified48. These genes appear to be differentially expressed in buds of poplar during the dormancy cycle (Rinne et al. unpublished). Some of them are orthologs of Arabidopsis seed oleosins, while others are somewhat closer to pollen oleosins. This similarity is interesting as the cells that constitute pollen and seeds arise from apex cells. Notably, all three of them also have the capacity to withstand extremes of desiccation, rehydration, heating and cooling for extended periods. The fact that oleosins enhance freezing tolerance in Arabidopsis7 suggests that LBs may also play such a role in pollen and overwintering apices. In seeds, ABA and osmotic stress upregulate LB caleosin19 and promote seed dormancy.72 In view of the similarities in LB regulation in apices and seeds, it seems plausible that the LBs in apices also possess caleosins, and possibly a number of other as yet unidentified proteins (Fig. 1E).
Figure 1.
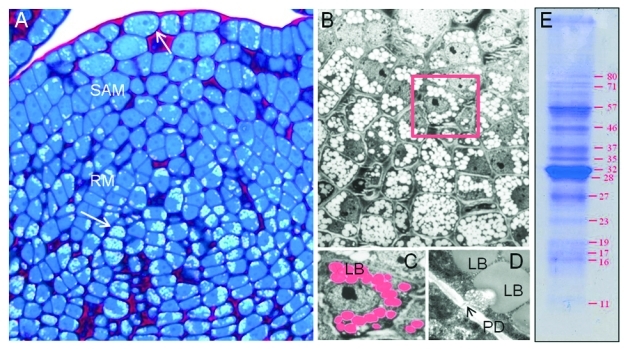
During short day (SD) exposure, apices of poplar accumulate numerous lipid bodies (LBs). (A). Light microscopy photo of a dormant shoot apex after 8 weeks of SD and 4 weeks of chilling. Arrows indicate lipid bodies. (B) Electron microscopy photo of dormant shoot apex after 8 weeks of SD. Boxed area is enlarged in c. (C) Single cell showing crowding LBs in pseudo color. (D). Lipid body-PD interaction during chilling. (E) Electrophoretic separation of proteins in the LB fraction isolated from apices after 8–10 weeks of SD.
Proteomic analyzes in combination with in situ immunolocalization studies indicated that LBs in birch contain 1,3-β-glucanases,65 also referred to as GH17 family proteins.67 These enzymes are crucial in releasing bud dormancy by reopening symplasmic paths that become obstructed by callose deposition during dormancy onset in early autumn.65,67,69 Also in poplar, a number of GH17 proteins are upregulated in the apex under SD concomitant with the production of LBs.67 Subsequent chilling during late autumn or early winter releases dormancy involving a GH17-based cellular mechanism.65,67 Central in this mechanism is that chilling induces displacement of LBs in apex cells to the PM in close proximity of PD, where the GH17 enzymes accumulate at both ends of the PD, resulting in the hydrolysis of PD-associated callosic sphincters (Fig. 2).65,67
Figure 2.
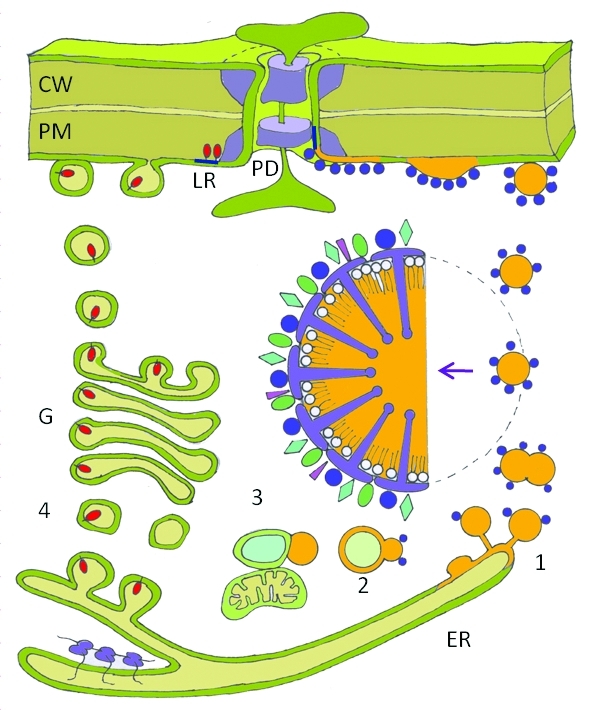
Speculative model depicting lipid bodies as vehicles for protein delivery to plasmodesmata in dormant buds. (1) Triacylglycerides (TAGs) are synthesized in the endoplasmic reticulum (ER), deposited within the ER membrane, where they diffuse to a budding site at a specialized distal part of the smooth ER to be released into the cytosol as a lipid body (LB). The LBs move to the PM to fuse with the cytoplasmic leaflet, from where LB proteins are recruited to microdomains or lipid rafts (blue) that are resident to the plasmodesmal (PD) channel. Although only the delivery of the LB-associated GH17 enzyme (purple) is depicted, LBs in poplar also contain the structural protein oleosin, probably caleosin and steroleosin, as well as a number of peripherally attached proteins (arrow; detailed cartoon of LB). (2) Some of the LBs target vacuoles for degradation. (3) Other LBs associate with glyoxysomes and mitochondria for lipolysis and subsequent cytoplasmic gluconeogenesis. (4) GPI-anchored GH17 enzymes (red) move via the Golgi excretion pathway (G) in vesicles to the PM where, after vesicle-PM fusion and release to the cell wall (CW), they are anchored to lipid rafts (LR, blue) at the extracellular leaflet of the PM and recruited to the PD neck.
In some other post-embryonic tissues LBs also need to be displaced in order to become effective. For example, in hydrating pollen, LBs gather near the germinative aperture, where the pollen tube emerges, and subsequently migrate into the forming pollen tube assisted by ER strands and cytoplasmic streaming.73,74 Another example is tapetum, a tissue of just one cell layer thick which encloses the microspores in the anther locule and controls their maturation. Tapetum cells express nine oleosin genes, the proteins of which accumulate in tapetosomes, a novel organelle type containing LBs.2 Tapetum cells are specialized in secretion, and during pollen maturation they release oleosins, but not TAGs, to the extracellular space to coat the surface of the pollen grains.2,75,76 Interestingly, the C-terminal extension part of oleosin is suggested to assist in the penetration of the lysing tapetum wall,2 which would be a novel oleosin function. As pointed out by Hsieh and Huang,12 this situation in tapetum reverses the importance of the LB components when compared with seeds. In seed LBs, the TAGs are the prime ingredient for physiological function while the oleosins are accessories. In contrast, in tapetosomes oleosins may be the main element for physiological function while TAGs are accessories.
In the perennial apex, the LB-associated GH17 proteins may act in a complementary fashion and in concert with GPI-lipid anchored GH17 proteins. As GPI-anchored proteins are thought to be tethered via their GPI-anchor to the outer PM leaflet,77 it seems plausible that the GPI-anchored GH17 protein AtBG-pap78,79 is delivered by secretory vesicles, via the Golgi system (Fig. 2).78-80 Lipid microdomains (35–70 nm)77 at the PM, often referred to as lipid rafts81 or membrane rafts77 might act as sorting devices for subsequent delivery to the PD.77,81 Since proteins might also anchor themselves at the cytoplasmic face of the PM, as evident from the clustered localization of the raft marker remorin,77 it cannot be excluded that some GH17 proteins are collected by microdomains at the cytoplasmic leaflet of the PM. While GPI-anchored GH17 proteins localize at the PM in a punctate pattern,67,78 LB-associated GH17 members appear to localize at the PM in sandwich-like patches at juxtaposed cells, as shown in leaves.67 This presumably reflects delivery by LBs that unload their cargo into a relatively wide area of the PM (Fig. 2), as the narrowness of the PD might preclude precise docking of LBs. Given the juxtaposed depositions it seems plausible that LBs dock to PM areas that are closely associated with PD, and that LB-associated GH17s from their collectively diffuse to PD,67 to be integrated in PD-resident microdomains or adhesion sites.77,81 It might be speculated that in the same way other peripherally associated LB proteins might be delivered to the PM, from where they diffuse to the PD to participate in the regulation of cell-to-cell transport (Fig. 2). Such lateral diffusion to PD has been observed in case of remorin, which also resides at the cytoplasmic leaflet of the PM.77
In chilling-induced dormancy release, LB associated GH17s might be of major importance to re-establish functional symplasmic path in the apex, including PD and young sieve tubes. Although applied gibberellic acid 4 (GA4) can bypass the chilling requirement by upregulating GH17s that are not LB-associated, the natural situation is based on mechanism activated by chilling.67 Chilling upregulates LB-associated GH17s and displaces the LBs toward the PM and PD.67 This underscores the central role LBs might have in re-establishing cell-cell signaling channels in the dormant apex. As in the apex, GA and GH17 enzymes also play a role in dormancy release in seeds.72 Since LBs in dehydrated seeds also align with the PM,83 it seems plausible that a similar mechanism as described for the dormant apex is operating in seeds.
Outlook: expansion of the paradigm
Recent studies have challenged the mundane view that LBs function only as inert lipid stores in seeds.6,14,53 First, although seed LBs are organelles that function in lipid storage, they appear to be far more dynamic than hitherto acknowledged.4,6,53 Second, the occurrence of LBs is more wide spread than previously assumed. Despite their obvious value as a paradigm for LBs in general, seed LBs appear to be special versions of LBs which are adapted and optimized to serve seed germination and post-germinative growth. On the other hand, in post-embryonic tissues, including vegetative and floral parts as well as pollen, LBs might have assumed functions that are absent or less significant in seeds.6 For example, the shoot-specific LB-associated caleosin RD20 in Arabidopsis (see above) is proposed to function as a signaling hub in a network of stress response pathways.53 Indeed, it appears that in LBs of post-embryonic tissues the enzymes are more important, although present only in minute amounts. If so, this would ask for a widening of the concept that defines what constitutes the LB proteome. Many new non-structural candidates for a redefined LB proteome might emerge, particularly in post-embryonic tissues.
In apices, LBs might have assumed at least three distinct functions: (1) to provide energy and carbon for growth after bud burst, (2) to provide lipids for reconstitution of the PM, (3) to transport enzymes and signals that may hitch a ride on LBs to modify the PM, cell wall, and PD. As overexpression of LB-associated GH17 enzymes in leaves of Nicotiana benthamiana results in the targeting of eGFP-tagged LB-associated GH17s to PD, LBs might have a more general role in all post-embryonic tissues in targeting enzymes and signaling peptides to the PM and PD. This putative role might be complementary to that of the Golgi cargo delivery system.80 Considering that viral agents either exploit the secretory pathway to reach the PD, or move as a vRNA-movement protein raft via the ER toward the PD,78-80,82 it is conceivable that LBs also deliver a cargo of opportunistic molecules to the PM and PD.
To identify proteins that are peripherally associated with LBs, less stringent isolation methods may be beneficial. Among the unavoidable false positives resulting from cytoplasmic contamination, such proteins could be identified by using in situ immuno-localization techniques and transgenic approaches. In addition, the validity of protein data could be confirmed by use of transgenic plants that express tagged LB markers as well as tagged peripherally associated enzymes or signaling peptides. Eventually, functional genomic studies, by knocking out specific oleosins or by interfering with LB-PM association, should be used to confirm that LBs are necessary vehicles for the delivery of enzymes or signals. A recent finding shows that the LB fraction of Arabidopsis seeds contains an aquaporin belonging to the family of tonoplast intrinsic proteins (TIPs), which may suggest that LBs may interact with vacuoles. We have also observed such interactions in apices.67 Although known as water channels, the TIPs were able to facilitate glycerol transport when expressed in yeast,84 suggesting that they may promote water uptake as well as glycerol transport in dehydrated seeds. Thus, LBs might be important during lipolysis and germination in seeds as well as in bud break and apical growth in perennial species.
To summarize, LBs in plants, like those in animals85 and fungi86 might be multifunctional organelles, participating in a host of cellular processes including membrane trafficking, lipid-based signaling, sterol-homeostasis, nutrient sensing, and transport of enzymes, regulatory proteins and signaling molecules.6,19,42,53 In the near future it will be of great interest to establish to what degree the LB- and PD proteome80,87,88 will converge.
Acknowledgments
Work in our lab is financially supported by the Norwegian Research Council (NFR), FRIBIO project grant 192013.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/17639
References
- 1.Pack DA. Dispersion of lipoids. Bot Gaz. 1925;79:334–8. doi: 10.1086/333482. [DOI] [Google Scholar]
- 2.Huang AHC. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996;110:1055–61. doi: 10.1104/pp.110.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Napier JA, Stobart AK, Shew PR. The structure and biogenesis of plant oil bodies: the role of the ER membrane and the oleosin class of proteins. Plant Mol Biol. 1996;31:945–56. doi: 10.1007/BF00040714. [DOI] [PubMed] [Google Scholar]
- 4.Murphy DJ. Biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/S0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 5.Tzen JTC, Peng C-C, Cheng D-J, Chen ECF, Chiu JMH. A new method for seed oil body purification and examination of oil body integrity following germination. J Biochem. 1997;121:762–8. doi: 10.1093/oxfordjournals.jbchem.a021651. [DOI] [PubMed] [Google Scholar]
- 6.Murphy DJ, ed. Plant lipids, biology, utilisation and manipulation. Blackwells, Oxford, 2004. [Google Scholar]
- 7.Jolivet P, Roux E, d’Andrea S, Davanture M, Negroni L, Zivy M, et al. Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem. 2004;42:501–9. doi: 10.1016/j.plaphy.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Frey-Wyssling A, Grieshaber E, Muhlethaler K. Origin of spherosomes in plant cells. J Ultrastruct Res. 1963;8:506–16. doi: 10.1016/S0022-5320(63)80052-0. [DOI] [Google Scholar]
- 9.Wanner G, Formanek H, Theimer RR. The ontogeny of lipid bodies (spherosomes) in plant cells. Planta. 1981;151:109–23. doi: 10.1007/BF00387812. [DOI] [PubMed] [Google Scholar]
- 10.Herman EM. Immunogold-localization and synthesis of an oil-body membrane protein in developing soybean seeds. Planta. 1987;172:336–45. doi: 10.1007/BF00398662. [DOI] [PubMed] [Google Scholar]
- 11.Yatsu LY, Jacks TJ. Spherosome membranes. Plant Physiol. 1972;49:937–43. doi: 10.1104/pp.49.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh K, Huang AHC. Endoplasmic reticulum, oleosins and oils in seeds and tapetum cells. Plant Physiol. 2004;136:3427–34. doi: 10.1104/pp.104.051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarmiento C, Ross JHE, Herman E, Murphy DJ. Expression and subcellular targeting of a soybean oleosin in transgenic rapeseed. Implications for the mechanism of oil-body formation in seeds. Plant J. 1997;11:783–96. doi: 10.1046/j.1365-313x.1997.11040783.x. [DOI] [PubMed] [Google Scholar]
- 14.Purkrtova Z, Jolivet P, Miquel M, Chardot T. Structure and function of seed lipid body-associated proteins. C R Biol. 2008;331:746–54. doi: 10.1016/j.crvi.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 15.Siloto RMP, Findlay K, Lopez-Villabos A, Yeung EC, Nykiforuk CL, Moloney MM. The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell. 2006;18:1961–74. doi: 10.1105/tpc.106.041269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trelease RN. Biogenesis of glyoxisomes. Annu Rev Plant Physiol. 1984;35:321–47. doi: 10.1146/annurev.pp.35.060184.001541. [DOI] [Google Scholar]
- 17.Buchanan BB, Gruissem W, Jones RL, eds. Biochemistry and Molecular Biology of Plants. ASPB 2000. [Google Scholar]
- 18.Hayashi Y, Hayashi M, Hasyashi H, Hara-Nishimura I, Nishimura M. Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant. Protoplasma. 2001;218:83–94. doi: 10.1007/BF01288364. [DOI] [PubMed] [Google Scholar]
- 19.Frandsen GI, Mundy J, Tzen JTC. Oil bodies and their associated proteins, oleosins and caleosins. Physiol Plant. 2001;112:301–7. doi: 10.1034/j.1399-3054.2001.1120301.x. [DOI] [PubMed] [Google Scholar]
- 20.Slack CR, Bertaud WS, Shaw BD, Holland R, Browse J, Wright H. Some studies of the composition and surface of oil bodies from the seed cotyledons of safflower and linseed. Biochem J. 1980;190:551–61. doi: 10.1042/bj1900551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang AHC. Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:177–200. doi: 10.1146/annurev.pp.43.060192.001141. [DOI] [Google Scholar]
- 22.Galili G, Sengupta-Gopalan C, Ceriotti A. The endoplasmic reticulum of plant cells and its role in protein maturation and biogenesis of oil bodies. Plant Mol Biol. 1998;38:1–29. doi: 10.1023/A:1006011919671. [DOI] [PubMed] [Google Scholar]
- 23.Tzen JTC, Lai Y-K, Chan K-L, Huang AHC. Oleosin isoforms of high and low molecular weights are present in the oil bodies of diverse seed species. Plant Physiol. 1990;94:1282–9. doi: 10.1104/pp.94.3.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu RD, Huang AHC. Oleosin KD 18 on the surface of oil bodies in maize. Genomic and cDNA sequences, and the deduced protein structure. J Biol Chem. 1990;265:2238–43. [PubMed] [Google Scholar]
- 25.Kim HU, Hsieh K, Ratnayake C, Huang AH. A novel group of oleosins is present inside the pollen of Arabidopsis. J Biol Chem. 2002;277:22677–84. doi: 10.1074/jbc.M109298200. [DOI] [PubMed] [Google Scholar]
- 26.Tai SSK, Chen MCM, Peng C-C, Tzen JTC. Gene family of oleosin isoforms and their structural stabilization in sesame seed oil bodies. Biosci Biotechnol Biochem. 2002;66:2146–53. doi: 10.1271/bbb.66.2146. [DOI] [PubMed] [Google Scholar]
- 27.Keddie JS, Tsiantis M, Piffanelli P, Cella R, Hatzopoulos P, Murphy DJ. A seed-specific Brassica napus oleosin promoter interacts with a G-box-specific protein and may be bi-directional. Plant Mol Biol. 1994;24:327–40. doi: 10.1007/BF00020171. [DOI] [PubMed] [Google Scholar]
- 28.Guilloteau M, Laloi M, Blais D, Crouzillat D, McCarthy J. Oil bodies in Theobroma cacao seeds: cloning and characterization of cDNA encoding the 15.8 and 16.9 kDa oleosins. Plant Sci. 2003;164:597–606. doi: 10.1016/S0168-9452(03)00011-6. [DOI] [Google Scholar]
- 29.Simkin AJ, Qian T, Caillet V, Michoux F, Ben Amor M, Lin C, et al. Oleosin gene family of Coffea canephora: quantitative expression analysis of five oleosin genes in developing and germinating coffee grain. J Plant Physiol. 2006;163:691–708. doi: 10.1016/j.jplph.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Wickner WT, Lodish HF. Multiple mechanisms of protein insertion into and across membranes. Science. 1985;230:400–7. doi: 10.1126/science.4048938. [DOI] [PubMed] [Google Scholar]
- 31.Abell BM, Holbrook LA, Albenes M, Murphy DJ, Hills MJ, Moloney MM. Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell. 1997;9:1481–93. doi: 10.1105/tpc.9.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lacey DJ, Wellner N, Beaudoin F, Napier JA, Shewry PR. Secondary structure of oleosins in oil bodies isolated from seeds of safflower (Catharantus tintosius L.) and sunflower (Helianthus annuus L.) Biochem J. 1998;334:469–77. doi: 10.1042/bj3340469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee K, Bih F-Y, Learn G, Ting JTL, Selles C, Huang AHC. Oleosins in the gametophytes of Pinus and Brassica and their phylogenetic relationship with those of various species. Planta. 1994;193:461–9. doi: 10.1007/BF00201827. [DOI] [PubMed] [Google Scholar]
- 34.Li M, Murphy DJ, Lee K-HK, Wilson R, Smith LJ, Clark DC, et al. Purification and structural characterization of the central hydrophobic domain of oleosin. J Biol Chem. 2002;277:37888–95. doi: 10.1074/jbc.M202721200. [DOI] [PubMed] [Google Scholar]
- 35.Cummins I, Hills MJ, Ross JH, Hobbs DH, Watson MD, Murphy DJ. Differential, temporal and spatial expression of genes involved in storage oil and oleosin accumulation in developing rapeseed embryos: implications for the role of oleosins and the mechanisms of oil-body formation. Plant Mol Biol. 1993;23:1015–27. doi: 10.1007/BF00021816. [DOI] [PubMed] [Google Scholar]
- 36.Chen MCM, Chyan CL, Lee TTT, Huang HC, Tzen JTC. Constitution of stable artificial oil bodies with triacylglycerol, phospholipid, and oleosin. J Agric Food Chem. 2004;52:3982–7. doi: 10.1021/jf035533g. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt MA, Herman EM. Suppression of soybean oleosin produces micro-oil bodies that aggregate into oil body/ER complexes. Mol Plant. 2008;1:910–24. doi: 10.1093/mp/ssn049. [DOI] [PubMed] [Google Scholar]
- 38.Vance VB, Huang AHC. The major protein from lipid bodies of maize: characterization and structure based on cDNA cloning. J Biol Chem. 1987;262:11275–9. [PubMed] [Google Scholar]
- 39.Tzen J, Cao YZ, Laurent P, Ratnayake C, Huang AHC. Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol. 1993;101:267–76. doi: 10.1104/pp.101.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capuano F, Beaudoin F, Napier JA, Shewry PR. Properties and exploitation of oleosins. Biotechnol Adv. 2007;25:203–6. doi: 10.1016/j.biotechadv.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Ross JHE, Sanchez J, Millan F, Murphy DJ. Differential presence of oleosins in oleogenic seed and mesocarp tissues in olive (Olea europea) and avocado (Persea americana) Plant Sci. 1993;93:203–10. doi: 10.1016/0168-9452(93)90050-A. [DOI] [Google Scholar]
- 42.Poxleitner M, Rogers SW, Samuels AL, Browse J, Rogers JC. A role for caleosine in degradation of oil-body storage lipid during seed germination. Plant J. 2006;47:917–33. doi: 10.1111/j.1365-313X.2006.02845.x. [DOI] [PubMed] [Google Scholar]
- 43.Hsiao ESL, Tzen JTC. Ubiquitination of oleosin-H and caleosin in sesame oil bodies after seed germination. Plant Physiol Biochem. 2011;49:77–81. doi: 10.1016/j.plaphy.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Naested H, Frandsen GI, Jauh G-Y, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, et al. Caleosins. Ca2+-binding proteins associated with lipid bodies. Plant Mol Biol. 2000;44:463–76. doi: 10.1023/A:1026564411918. [DOI] [PubMed] [Google Scholar]
- 45.Thoyts PJ, Millichip MI, Stobart AK, Griffiths WT, Shewry PR, Napier JA. Expression and in vitro targeting of a sunflower olesosin. Plant Mol Biol. 1995;29:403–10. doi: 10.1007/BF00043664. [DOI] [PubMed] [Google Scholar]
- 46.van Rooijen GJH, Moloney MM. Plant seed oil-bodies as carriers for foreign proteins. Biotechnol (NY) 1995;13:72–7. doi: 10.1038/nbt0195-72. [DOI] [PubMed] [Google Scholar]
- 47.Jiang P-L, Jauh G-Y, Wang C-S, Tzen JTC. A unique caleosin in oil bodies of lily pollen. Plant Cell Physiol. 2008;49:1390–5. doi: 10.1093/pcp/pcn103. [DOI] [PubMed] [Google Scholar]
- 48.Huang C-Y, Chung C-I, Lin Y-C, Hsing Y-IC, Huang AHC. Oil bodies and oleosins in Physcomitrella possess characteristics representative of early trends in evolution. Plant Physiol. 2009;150:1192–203. doi: 10.1104/pp.109.138123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen JCF, Tsai CCY, Tzen JTC. Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of seed plants. Plant Cell Physiol. 1999;40:1079–86. doi: 10.1093/oxfordjournals.pcp.a029490. [DOI] [PubMed] [Google Scholar]
- 50.Lin L-J, Sorgan SK, Peng C-C, Tzen JTC. Steroleosin, a sterole-binding dehydrogenase in seed oil bodies. Plant Physiol. 2002;128:1200–11. doi: 10.1104/pp.010982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signaling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 52.Hanano A, Burcklen M, Flenet M, Ivancich A, Louwagie M, Garin J, et al. Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J Biol Chem. 2006;281:33140–51. doi: 10.1074/jbc.M605395200. [DOI] [PubMed] [Google Scholar]
- 53.Aubert Y, Vile D, Pervent M, Aldon D, Ranty B, Simonneau T. RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2010;51:1975–87. doi: 10.1093/pcp/pcq155. [DOI] [PubMed] [Google Scholar]
- 54.Chen ECF, Tai SSK, Peng CC, Tzen JTC. Identification of three novel unique proteins in seed oil bodies of sesame. Plant Cell Physiol. 1998;39:935–41. doi: 10.1093/oxfordjournals.pcp.a029457. [DOI] [PubMed] [Google Scholar]
- 55.Tzen JTC, Wang MMV, Chen JCF, Lin LJ, Chen MCM. Seed oil body proteins: oleosin, caleosin, and steroleosin. Curr Topics Biochem Res. 2003;5:133–9. [Google Scholar]
- 56.Li F, Asami T, Wu X, Tsang EW, Cutler AJ. A putative hydroxysteroid dehydrogenase involved in regulating plant growth and development. Plant Physiol. 2007;145:87–97. doi: 10.1104/pp.107.100560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hause B, Weichert H, Hohne M, Kindl H, Feussner I. Expression of cucumber lipid-body lipoxygenase in transgenic tobacco: lipid-body lipoxygenase is correctly targeted to seed lipid bodies. Planta. 2000;210:708–14. doi: 10.1007/s004250050671. [DOI] [PubMed] [Google Scholar]
- 58.Eastmond PJ. Cloning and characterization of the acid lipase from castor beans. J Biol Chem. 2004;279:45540–5. doi: 10.1074/jbc.M408686200. [DOI] [PubMed] [Google Scholar]
- 59.Eastmond PJ. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell. 2006;18:665–75. doi: 10.1105/tpc.105.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.May C, Hohne M, Gnau P, Schwennesen K, Kindl H. The N-terminal beta-barrel structure of lipid body lipoxygenase mediates its binding to liposomes and lipid bodies. Eur J Biochem. 2000;267:1100–9. doi: 10.1046/j.1432-1327.2000.01105.x. [DOI] [PubMed] [Google Scholar]
- 61.Holk A, Rietz S, Zahn M, Quader H, Scherer GF. Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiol. 2002;130:90–101. doi: 10.1104/pp.006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoppe A, Theimer M. Degradation of oil bodies isolated from cotyledons during germination. J Plant Physiol. 1997;151:471–8. [Google Scholar]
- 63.Tzen JT, Lie GC, Huang AH. Characterization of the charged components and their topology on the surface of plant seed oil bodies. J Biol Chem. 1992;267:15626–34. [PubMed] [Google Scholar]
- 64.Rinne PLH, Kaikuranta PLM, van der Plas LHW, van der Schoot C. Dehydrins in cold-acclimated apices of birch (Betula pubescens Ehrh.): production, localization and potential role in rescuing enzyme function during dehydration. Planta. 1999;209:377–88. doi: 10.1007/s004250050740. [DOI] [PubMed] [Google Scholar]
- 65.Rinne PLH, Kaikuranta PM, van der Schoot C. The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J. 2001;26:249–64. doi: 10.1046/j.1365-313X.2001.01022.x. [DOI] [PubMed] [Google Scholar]
- 66.Rinne PLH, van der Schoot C. Plasmodesmata at the cross roads between development, dormancy, and defense. Can J Bot. 2003;81:1182–97. doi: 10.1139/b03-123. [DOI] [Google Scholar]
- 67.Rinne PLH, Welling A, Vahala J, Ripel L, Ruonala R, Kagasjärvi K, et al. Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-β-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell. 2011;23:130–46. doi: 10.1105/tpc.110.081307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rohde A, Prinsen E, de Rycke R, Engler G, van Montagu M, Boerjan W. PtABI34 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell. 2002;14:1885–901. doi: 10.1105/tpc.003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ruonala R, Rinne PLH, Kangasjärvi J, van der Schoot C. CENL1 expression in the rib meristem affects stem elongation and the transition to dormancy in Populus. Plant Cell. 2008;20:59–74. doi: 10.1105/tpc.107.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van der Schoot C, Rinne PLH. Dormancy cycling at the shoot apical meristem: transitioning between self-organization and self-arrest. Plant Sci. 2011;180:120–31. doi: 10.1016/j.plantsci.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 71.Shimada TL, Shimada H, Takahashi H, Fukao Y, Haranishimura I. A novel role of oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 2008;55:798–809. doi: 10.1111/j.1365-313X.2008.03553.x. [DOI] [PubMed] [Google Scholar]
- 72.Leubner-Metzger G. Functions and regulation of β-1,3-glucanases during seed germination, dormancy release and after-ripening. Seed Sci Res. 2003;13:17–34. doi: 10.1079/SSR2002121. [DOI] [Google Scholar]
- 73.Rodríguez-García MI, M’rani-Alaoui M, Fernández MC. Behaviour of storage lipids during development and germination of olive (Olea europaea L.) pollen. Protoplasma. 2003;221:237–44. doi: 10.1007/s00709-002-0076-x. [DOI] [PubMed] [Google Scholar]
- 74.Zienkiewicz K, Castro AJ, de Dios Alché J, Zienkiewicz A, Suárez C, Rodriguez-Garcia MI. Identification and localization of a caleosin in olive (Olea europea L.) pollen during in vitro germination. J Exp Bot. 2010;61:1537–46. doi: 10.1093/jxb/erq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu SS, Platt KA, Ratnayake C, Wang TW, Ting JT, Hung AH. Isolation and characterisation of novel neutral-lipid-containing organelles and globuli-filled plastids from Brassica napus tapetum. Proc Natl Acad Sci USA. 1997;94:12711–6. doi: 10.1073/pnas.94.23.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsieh K, Huang AH. Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-like flavonoids and alkanes for delivery to the pollen surface. Plant Cell. 2007;19:582–96. doi: 10.1105/tpc.106.049049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mongrand S, Staislas T, Bayer EMF, Lherminier J, Simin-Plas F. Membrane rafts in plant cells. Trends Plant Sci. 2010;15:656–63. doi: 10.1016/j.tplants.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 78.Levy A, Erlanger M, Rosenthal M, Epel BL. A plasmodesmata-associated β-1-3-glucanase in Arabidopsis. Plant J. 2007;49:669–82. doi: 10.1111/j.1365-313X.2006.02986.x. [DOI] [PubMed] [Google Scholar]
- 79.Levy A, Guenoune-Gelbart D, Epel BL. β-1-3-Glucanases. Plasmodesmal gate keepers for intercellular communication. Plant Signal Behav. 2007;2:404–7. doi: 10.4161/psb.2.5.4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oparka KJ. Getting the message across: how do plant cells exchange macromolecular complexes? Trends Plant Sci. 2004;9:33–41. doi: 10.1016/j.tplants.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 81.Tilsner J, Amari K, Torrance L. Plasmodesmata viewed as specialised membrane adhesion sites. Protoplasma. 2011;248:39–60. doi: 10.1007/s00709-010-0217-6. [DOI] [PubMed] [Google Scholar]
- 82.Epel BL. Plant viruses spread by diffusion on ER-associated movement-protein-rafts through plasmodesmata gated by viral induced host β-1-3-glucanases. Semin Cell Dev Biol. 2009;20:1074–81. doi: 10.1016/j.semcdb.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 83.Cordova-Tellez L, Burris JS. Alignment of lipid bodies along the plasma membrane during the acquisition of dessication tolerance in maize seed. Crop Sci. 1982;42:1982–8. doi: 10.2135/cropsci2002.1982. [DOI] [Google Scholar]
- 84.Weig AR, Jakob C. Functional identification of the glycerol permease activity of Arabidopsis thaliana NLM1 and NML2 proteins by heterologous expression in Saccharomyces cerevisiae. FEBS Lett. 2000;481:293–8. doi: 10.1016/S0014-5793(00)02027-5. [DOI] [PubMed] [Google Scholar]
- 85.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–40. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kamisaka Y, Noda N, Yamaoka M. Appearance of smaller lipid bodies and protein kinase activation in the lipid body fraction are induced by an increase in the nitrogen source in the Mortierella fungus. J Biochem. 2004;135:269–76. doi: 10.1093/jb/mvh032. [DOI] [PubMed] [Google Scholar]
- 87.Faulkner C, Maule A. Opportunities and successes in the research for plasmodesmal proteins. Protoplasma. 2011;248:27–38. doi: 10.1007/s00709-010-0213-x. [DOI] [PubMed] [Google Scholar]
- 88.Fernandez-Calvino L, Faulkner C, Walshaw J, Saalbach G, Bayer E, Benitez-Alfonso Y, et al. Arabidopsis plasmodesmal proteome. PLoS One. 2011;6:e18880. doi: 10.1371/journal.pone.0018880. [DOI] [PMC free article] [PubMed] [Google Scholar]


