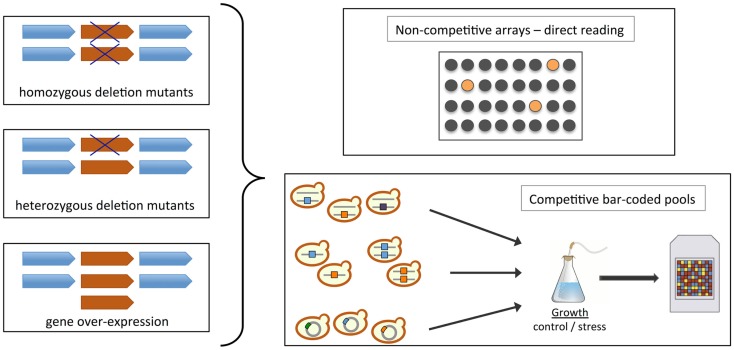
Figure 2.
Construction and screening of yeast collections. Schematic representation of methodologies and cell libraries available for chemogenomics testing in S. cerevisiae (homozygous or haploid deletion – gene dosage 0%, heterozygous deletion – gene dosage 50%, and overexpression – gene dosage > 100%; see Section “Functional Toxicogenomics using Yeast Gene Deletion Collections”; Auerbach et al., 2005; Hoon et al., 2008b; Wuster and Madan Babu, 2008; North and Vulpe, 2010; Smith et al., 2010). The fitness of strains upon chemical treatment is usually assessed in non-competitive arrays or in competitive bar-coded pools. In the first case, the toxicant can be added to a well plate and each mutant occupies a separate well; the effects are observed directly by comparison with wild-type strain fitness. In the second case, the screen is executed in a pooled format where uniquely tagged (“bar-coded”) strains are grown together in the presence of a toxicant. Fitness is assessed by determining the abundance of the different mutant strains using microarrays coupled with a PCR strategy that amplifies the molecular bar-codes associated with each mutant. Strain depletion in the toxicant-treated pool indicates chemical hypersensitivity.