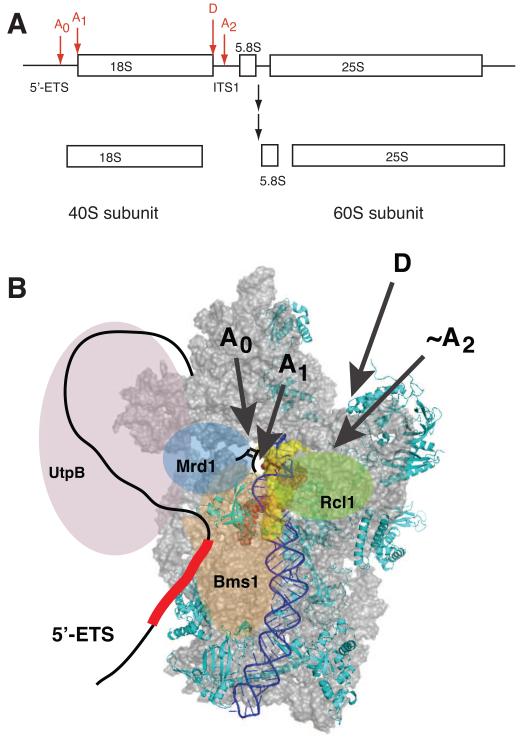
Figure 3.
(A) pre-rRNA precursor and cleavage sites for 18S rRNA production. (B) Structural model of early pre-40S ribosomes, viewed from the subunit interface. Only Rps required for early steps are depicted [*27]. H44 is highlighted in blue. Nucleotides that form base pairs with U3 snoRNA and ITS1 are shown in red or yellow spacefill, respectively. In early pre-ribosomes the top of H44, which otherwise covers the U3-interacting regions, is disrupted and might be dislocated to allow access to U3 snoRNA [*45]. The nucleotides that form base pairs with the yellow region in H44 are directly downstream of cleavage site A2 [*45], which allows the rough location of that site. The 5′- and 3′-ends of 18S rRNA (sites A1 and D) point toward the back, as indicated with arrows. Bms1 and Rcl1 are positioned where Tsr1 binds, and near the A2 site, respectively. U3 snoRNA binds the red regions in 18S rRNA, as well as regions in the 5′-ETS, schematically depicted as a red box, and for clarity is not explicitly shown. Mrd1 is positioned to bind near the U3 binding site, as well as near UtpB, in keeping with interactions with both ([58], see Figure 2). The UtpB complex, shown in lavender, forms interactions with the U3/Mpp10 complex, and Enp1 and Ltv1 [36,37,40,41,62], located at the back of the beak [**28,52,*53].