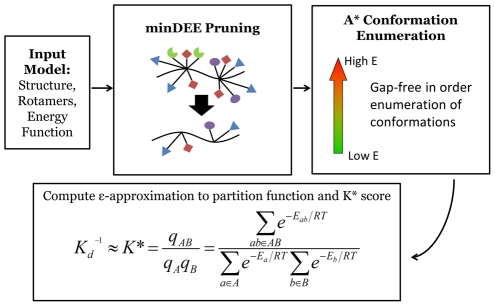
Figure 2. Overview of  Algorithm.
Algorithm.
he  algorithm searches over protein sequences and conformations to find the protein complexes with the best binding constant.
algorithm searches over protein sequences and conformations to find the protein complexes with the best binding constant.  takes an input model composed of an initial protein structure, a rotamer library to search over side-chain conformations, and an energy function to evaluate conformations. Minimization-aware DEE (minDEE) prunes rotamers that are not part of the lowest energy conformations for a given sequence. The remaining conformations from minDEE are enumerated in order of increasing energy lower bounds using A*. Finally, the conformations are Boltzmann-weighted and used to compute partition functions and ultimately a
takes an input model composed of an initial protein structure, a rotamer library to search over side-chain conformations, and an energy function to evaluate conformations. Minimization-aware DEE (minDEE) prunes rotamers that are not part of the lowest energy conformations for a given sequence. The remaining conformations from minDEE are enumerated in order of increasing energy lower bounds using A*. Finally, the conformations are Boltzmann-weighted and used to compute partition functions and ultimately a 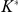 score for each sequence.
score for each sequence.