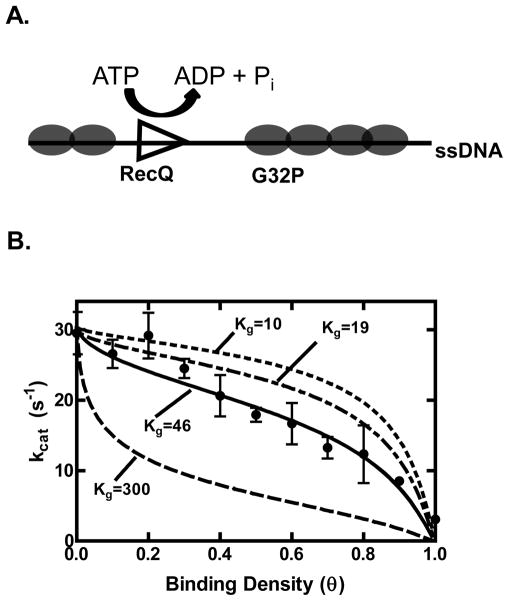
Figure 5. G32P inhibits the ATP hydrolysis activity of RecQ by creating short gaps of ssDNA.
(A) Diagram of the experiment. Increasing the binding density (θ) decreases the lengths of ssDNA available for RecQ binding and translocation. (B) The rate of ATP hydrolysis by RecQ (100 nM) stimulated by poly dT (2 μM) is plotted as a function of increasing concentrations of G32P (black circles). The data from θ = 0 to 0.9 were fit using equation 3 (solid line), and values of 32±2 s−1 and 46±11 nucleotides were obtained for Vf and Kg, respectively. Also shown are simulations of equation 3 with the value of Vf set to 32 s−1 and Kg set to either 10, 19 or 300 nucleotides (dashed lines). The turnover number for θ = 1 was obtained by adding G32P at 1 μM (3.75-fold in excess of the ssDNA).