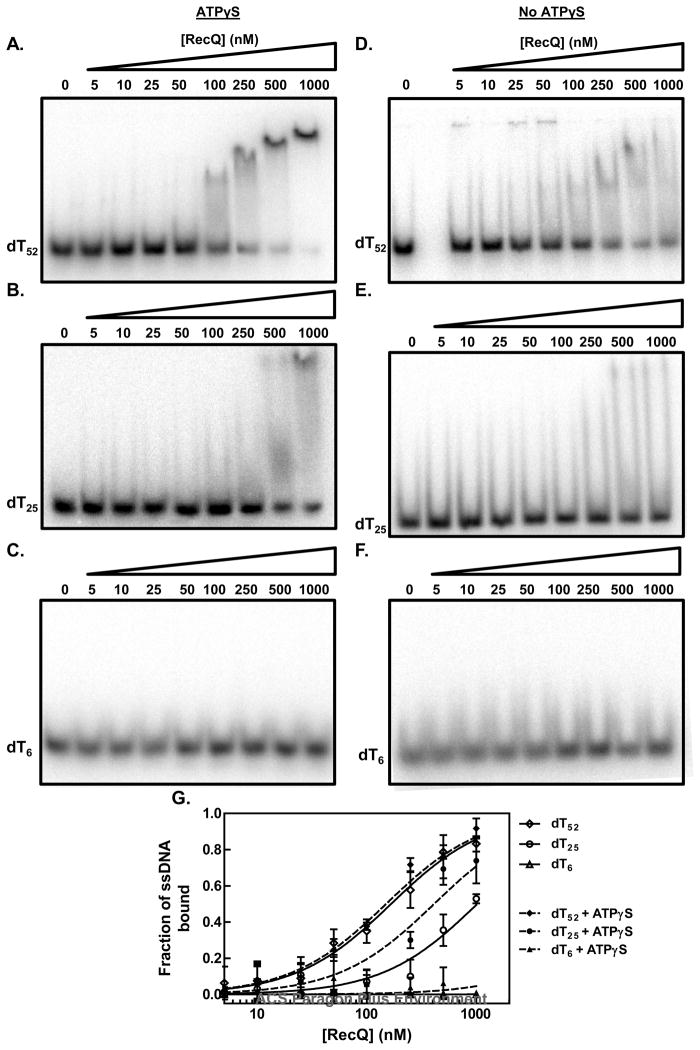
Figure 6. The apparent affinity of RecQ for ssDNA is dependent on the lattice length.
The apparent affinity of RecQ for oligonucleotides was assayed by using the electrophoretic mobility shift assay. (A–C) in the presence of ATPγS; (D–F) in the absence of any nucleotide cofactor; (G) quantification of the disappearance of free DNA relative to the intensity of DNA lane without RecQ: the absence of nucleotide cofactor (open symbols) or presence of ATPγS (filled symbols). The apparent binding constant, Kd, was determined for each DNA substrate by fitting the data to the equation for a one-site binding curve. The fitted curves are shown for each substrate in the absence of a nucleotide cofactor (solid lines) or presence of ATPγS (dashed lines).