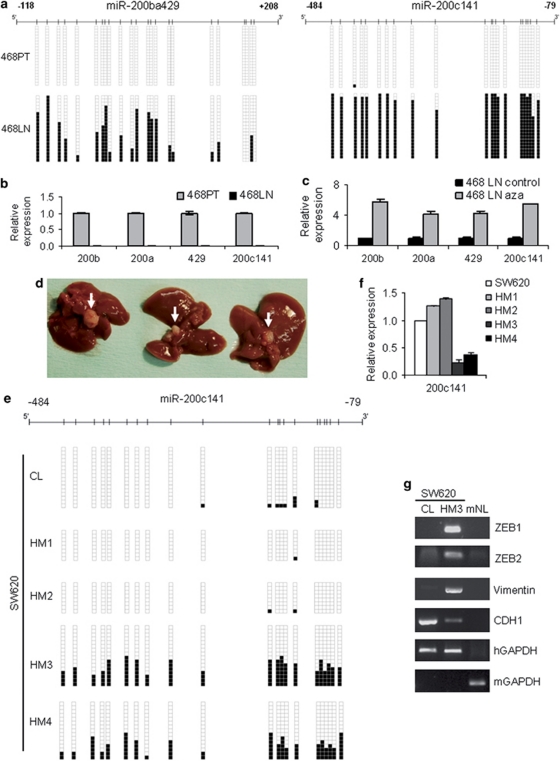
Figure 4.
Dynamic epigenetic silencing of the miRNA-200 family during in vivo metastasis formation. (a) CpG hypermethylation of both miR-200 clusters in the lung metastasis-derived 468LN cell line, in contrast to unmethylated status in the parental 468PT epithelial breast cancer cell line, analyzed by bisulfite genomic sequencing. In all, 20 single clones are represented for each cell line. Presence of unmethylated or methylated CpGs is indicated by white or black squares, respectively. (b) Significant reduction of miR-200 expression in metastatic 468LN cells (Student's t-test P<0.01). Data represent relative quantification of miRNAs, using the parental 468PT cell line as reference. (c) Restored expression of miR-200 in 468LN cells upon DNA-demethylating treatment with 5-aza-2′-deoxycytidine (aza), detected by qRT–PCR (Student's t-test P<0.05). (d) CpG island hypermethylation of miR-200c141 cluster during hepatic metastasis formation in mice. Spleen injection of 2 × 106 human colon cancer SW620 cells was able to generate hepatic metastases in 9 out of 15 mice after 8 weeks. Illustrative examples of metastases are shown (white arrows). (e) miR-200c141 CpG island methylation analysis of the hepatic metastases derived from SW620 shows hypermethylation of the cluster in two of them (HM3 and HM4), in contrast to the unmethylated status of the original SW620 cell line (CL). Results were obtained by bisulfite genomic sequencing analysis. Presence of unmethylated or methylated CpGs is indicated by white or black squares, respectively. (f) Significant decrease of pri-miR-200c141 in a CpG-hypermethylated hepatic metastasis (HM3) in comparison with the unmethylated metastases and original SW620 cell line (Student's t-test P<0.01). Data represent means±s.d. and were obtained by qRT–PCR. (g) Gain of expression of ZEB1, ZEB2 and Vimentin, and repression of E-cadherin in a miR-200c141-methylated hepatic metastasis (HM3). Human (hGAPDH) or mouse GAPDH (mGAPDH) were used as endogenous controls in semiquantitative RT–PCR. CL: original SW620 cell line; mNL: mouse normal liver.