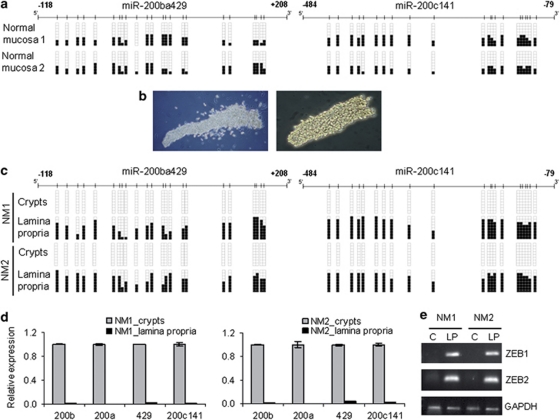
Figure 5.
Differential miR-200 CpG island methylation in epithelial and stromal cells of normal human colonic mucosa. (a) miR-200ba429 and miR-200c141 5′-CpG island methylation analyses by bisulfite genomic sequencing in primary total normal colon mucosa samples demonstrated a mixture of unmethylated and methylated alleles. Illustrative examples are shown. (b) Epithelial and stromal components of human colonic mucosa were mechanically isolated. Illustrative microscope photographs in conventional bright-field illumination (left panel) or under phase contrast (right panel) of epithelial colonic crypts are shown. (c) Lack of DNA methylation in miR-200 CpG islands in epithelial component (colonic crypts), but hypermethylation in the surrounded lamina propria-stromal cells derived from normal colonic mucosa samples (NM), studied by bisulfite genomic sequencing. Illustrative examples are shown. (d) High miR-200 expression levels in unmethylated colonic crypts and loss of expression in methylated lamina propria cells, evaluated by qRT–PCR of primary miRNAs (Student's t-test P<0.01). (e) Inhibition of ZEB1 and ZEB2 mRNA expression in miR-200 unmethylated crypts (c), in contrast to high expression in miR-200-methylated lamina propria (LP) cells. GAPDH was used as endogenous control in semiquantitative RT–PCRs.