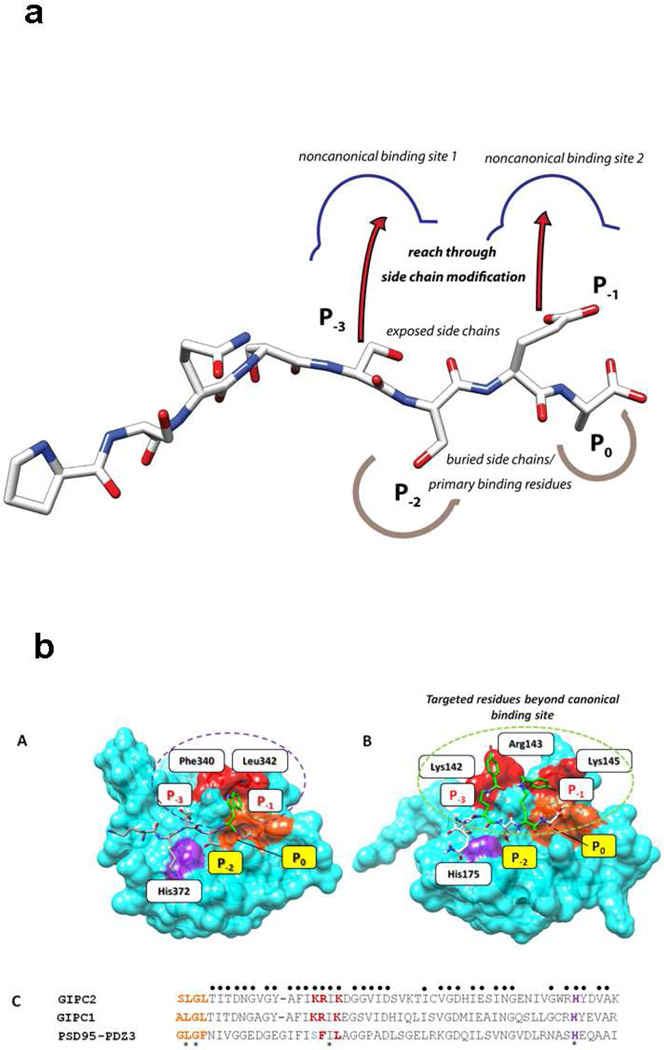
Figure 6. Design of the modified compounds based on the sequence of parent active compound, PSQSSSEA (CR1023,).
a. The P−1 and/or P−3 positions are replaced by Lys acylated with an organic acid (halogenated or non-halogenated). If only P−3 is modified, P−1 is Glu, if only P−1 position is modified, P−3 is Ser. b. Structure-based design rationale for halogenated benzoyl-modified ligands for GIPC PDZ domains, based on the complex between PDZ3 and KKETWV. Positions P0 and P−2 denote conserved primary binding determinants. (A) Key residues of PDZ3 domain binding interaction with the KKETWV are shown with conserved residues for canonical binding sites and the P−1 interaction site. (B) Expected binding mode of the pharmacophore region of CR1166 [QSK(4-bromobenzoyl)SK(4-bromobenzoyl)A] in the GIPC PDZ domain. The residues (Gly-Leu-Gly-Phe) of the carboxylate binding GLGF loop (of PDZ3) and its counterpart SLGL (in GIPC2) are colored in orange, and the His crucial for P−2 binding is shown in purple. Additional residues contacting ligands in both PDZ3 and GIPC PDZ complexes are red. The red surface represents the targeted region (P−3 and/or P−1) in PDZ proteins. In PDZ3 hydrophobic residues (Phe and Leu) line the cavity, in GIPC these are positively charged resides (Arg and Lys). In the design, either P−1 or P−3, or both, positions will be replaced by Lys acylated with halogenated benzoic acid. (C) Multiple sequence alignment of selected region of PDZ domains of GIPC2, RGS-GAIP interacting protein (GIPC1) and PSD-95-PDZ3. A sequence alignment of GIPC2, RGS-GAIP interacting protein (GIPC1) and PSD95-PDZ3 was performed with T-Coffee (www.ebi.ac.uk/t-coffee). The sequences were arranged according to structure-based alignment. The key residues identical in all three regions are marked with an asterisk, those identical to either GIPC PDZ domain regions are marked with a dot. The color scheme is as in A. The alignment scores for all three PDZ regions and for the two GIPC PDZ proteins are 81% and 98%, respectively. The amino acid sequences are as follows: GIPC2 chain A (PDB ID: 3GGE), RGS-GAIP (GIPC1) (accession AAC67550.1), and PSD95-PDZ3 (PDB ID: 1TP5). Either P−1 (Glu) or P−3 (Ser) or both positions are replaced by Lys acylated with organic acid (halogenated or non-halogenated). Structures were generated from Chem3D Pro program v11.0.1, AutoDock Tools 1.5.4 and rendered using UCSF Chimera v1.5.