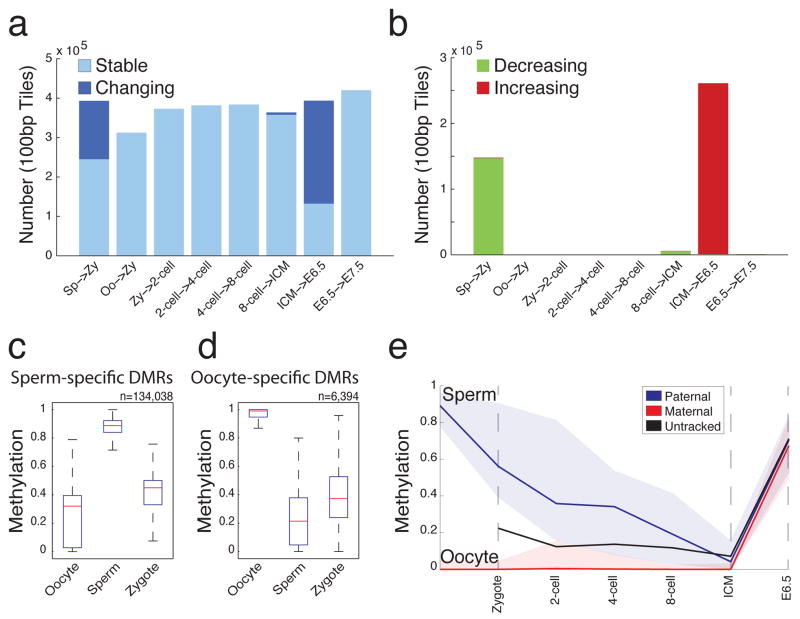
Figure 2. Major transitions in DNA methylation levels during early development.
a. 100bp tiles available for pairwise comparison across consecutive embryonic stages. Tiles that remain unchanged (stable) at the indicated transitions are shown in light blue. Tiles that change by greater than 0.2 and are significant by t-test are highlighted in dark blue.
b. 100bp tiles with increasing (red) or decreasing (green) methylation levels at each consecutive transition show that major transitions are largely unidirectional.
c. Boxplot of methylation levels for sperm-specific DMRs (n=134,038 tiles). Red line indicates the median, edges the 25th/75th percentile and whiskers the 2.5th/97.5th percentile.
d. Boxplot of methylation levels for oocyte-specific DMRs (n=6,394 tiles) as in (c).
e. 74 CpGs within sperm-specific DMR tiles (c) could be ascribed to paternal and maternal alleles and tracked across stages. Paternal CpG methylation values (blue line, median; colored space, 25th/75th percentile) exhibit marked decrease by the zygote stage while maternal CpG methylation (red line, median; colored space, 25th/75th percentile) remain unchanged. If untracked, these CpGs have an intermediate methylation value between those ascribed to a parent-of-origin (black line).