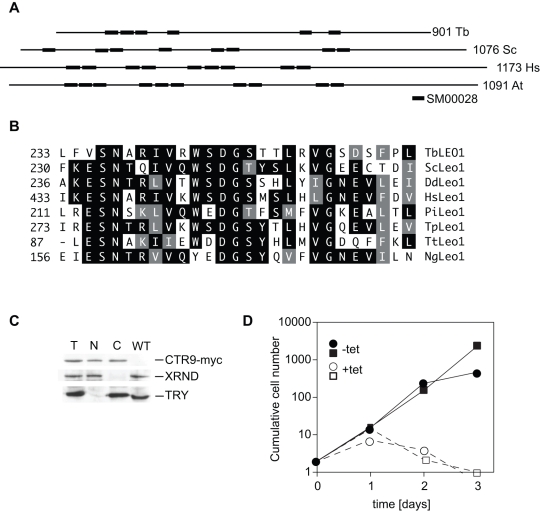
Figure 1. Properties of CTR9 complex components.
A. The locations of TPR repeats (SMART SM00028) in trypanosome (Tb), Saccharomyces cerevisiae (Sc), human (Hs) and Arabidopsis thaliana (At) Ctr9 homologues. The maps are taken from the Interpro scan web site. Domains are shown as thickened bars. B. The most conserved portion of the Leo1 domain. The alignment was made using the MegAlign portion of the DNAStar package; for details see Figure S3. Identical amino acids are underlain in black and functionally conserved amino acids are underlain in grey. Dd: Dictyostelium disoideum; Pi: Phytophthora infestans – Tp: Thalassiosira pseudonana; Tt: Tetrahymena thermophila; Ng: Naegleria gruberi. C. Myc-tagged CTR9 fractionates into both the nucleus and the cytoplasm. Cells expressing CTR9-myc were lysed with NP40, centrifuged, and the extracts subjected to Western blotting using antibodies to myc (Santa Cruz), XRND (nuclear marker) and trypanothione reductase (cytoplasmic marker). T: total; N: nuclear fraction; C: cytoplasmic fraction. D. Effect of RNAi against CTR9. Two separate bloodstream trypanosome lines (represented by different symbols) were incubated with or without 100 ng/ml tetracycline. The starting cell density was 2×105/ml and cultures were diluted to that level daily as required.