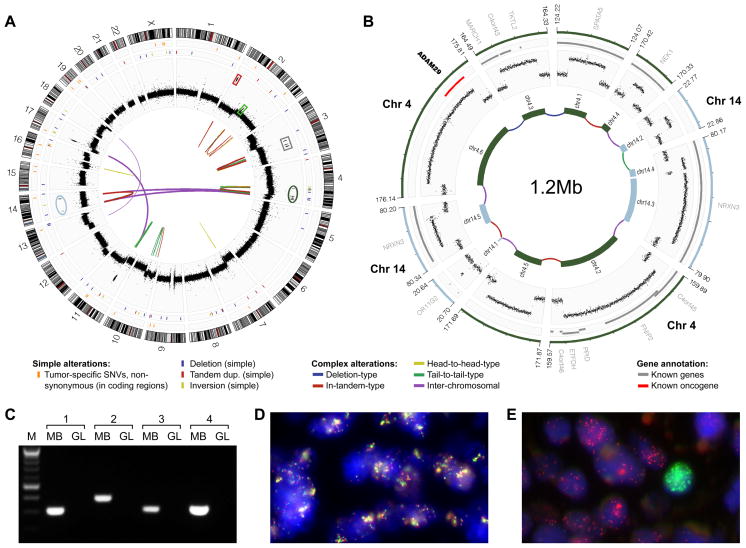
Figure 1.
Analysis of LFS-MB1 revealed catastrophic DNA rearrangements consistent with chromothripsis. (A) Genome-wide distribution of somatic DNA variants. Thin orange lines in outer-most panel are non-synonymous somatic SNVs; the next panel shows isolated genomic rearrangements. Read-depth plots (log2-ratio tumor vs. germline), indicating copy-number alterations, are in black. Connecting lines show complex large-scale (e.g., inter-chromosomal) rearrangements identified by paired-end mapping. (B) Inferred double minute chromosome structure (originating segments from chromosome 4 and 14 are highlighted in panel (A)). Genes are in gray (known cancer genes are in red). (C) PCR validation of inter-chromosomal rearrangements contributing to the inferred double minute chromosome. MB, medulloblastoma; GL, germline. (D) FISH validation of rearrangements contributing to double minute chromosome derived from chromosome 3 segments. Probes match to normally distal regions of chromosome 3 (RP11-553D4, red, and RP11-265F19, light green; see panel (A) and Figure S1). (E) Amplification of MYCN (red) and GLI2 (light green), not associated with chromothripsis (amplicon loci highlighted in panel (A) with red and light green boxes), was observed in distinct subpopulations of cells.