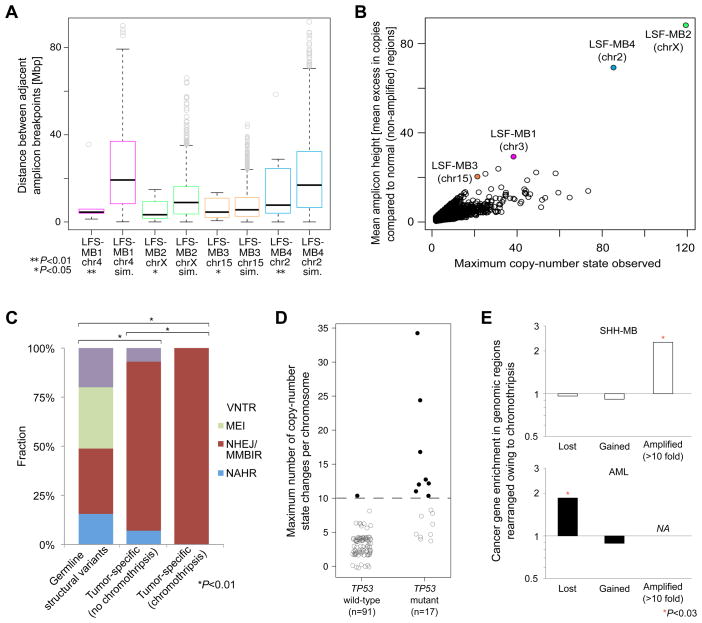
Figure 5.
Analysis of chromothripsis-associated DNA rearrangements in SHH-MB and AML. (A) Topographical clustering of amplified regions rearranged by chromothripsis. Sim., simulated amplicon distances (P-values are based on 1000 permutations). (B) Simulations of progressive rearrangements segregate from the actual data in terms of mean excess in copy-number compared to unaffected regions. (C) Rearrangement formation mechanisms analysis. Polymorphic genomic structural variants detected in the germline are shown for comparison. P-values, indicating significant differences between the distributions of inferred formation mechanisms, are based on Chi-square tests. VNTR, expansion or shrinkage of regions with variable number of tandem repeats; MEI, mobile element insertions; NAHR, non-allelic homologous recombination (other abbreviations: see main text). (D) Somatically acquired TP53 mutations are linked with the occurrence of chromothripsis in AML. Black filled circles: AMLs with chromothripsis. Gray open circles: AMLs without chromothripsis. Example copy-number profiles are available as Data S2, and a detailed summary of the AML data is in Table S5. (E) Cancer gene enrichment in association with chromothripsis in SHH-MBs, analyzed by deep sequencing, and AMLs, analyzed by SNP arrays. Regions hemizygously deleted (‘lost’), gained, and highly (>10-fold) amplified as a consequence of chromothripsis were separately analyzed. No genes displayed high-level amplification in AML in association with chromothripsis (‘NA’). ‘*’significant based on Fisher’s exact test.