Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) with exogenous cassette DNA containing the methicillin-resistant gene mecA (SCCmec) poses a problem as a drug-resistant bacterium responsible for hospital- and community-acquired infections. The frequency of MRSA detection has recently been increasing rapidly in Japan, and SCCmec has also been classified more diversely into types I–V. A rapid test is essential for early diagnosis and treatment of MRSA infections, but detection by conventional methods requires at least two days. The newly developed multiplex PCR lateral flow method allows specific amplification of femA to detect S. aureus, mecA to detect SCCmec, and kdpC to detect SCCmec type II; moreover, PCR products can be evaluated visually in about 3 h. In the present study, we developed a PCR lateral flow method for MRSA using this method and investigated its clinical usefulness in the detection of MRSA. The results showed a diagnostic concordance rate of 91.7% for MRSA and methicillin-susceptible S. aureus between bacteriological examination and PCR lateral flow, and a high level of specificity in PCR lateral flow. In addition, a higher detection rate for S. aureus using the same sample was observed for PCR lateral flow (70.2%) than for bacteriological tests (48.6%). The above results show that PCR lateral flow for MRSA detection has high sensitivity, specificity, and speed, and its clinical application as a method for early diagnosis of MRSA infections appears to be feasible.
KEY WORDS: multiplex PCR, lateral flow, MRSA, rapid genetic testing
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) was first reported in Britain in 1961 as Staphylococcus aureus with exogenous cassette DNA containing the methicillin-resistant gene mecA (SCCmec) [1]. MRSA is currently monitored as one of the major causes of hospital-acquired infections in Japan. However, the Centers for Disease Control and Prevention (CDC) also reported deaths due to community-acquired MRSA in 1999. MRSA also poses a problem as a drug-resistant bacterium responsible for community-acquired infections where there are no risk factors such as a history of hospitalization [2, 3]. The frequency with which MRSA is detected has recently been increasing rapidly in Japan, and SCCmec has also been classified more diversely into types I–V [4]. These findings demonstrate that the potential spread of MRSA among immunocompromised individuals could be a more serious threat than previously considered.
A rapid test method is therefore essential for early diagnosis and treatment of MRSA infections. However, detection of MRSA by standard bacteriological testing requires at least 2 days. Furthermore, recently investigated and developed methods such as the cefoxitin (CFX) susceptibility test, the PBP latex agglutination test, the PNA-FISH method, and real-time PCR require expensive special measuring instruments and also show low sensitivity and specificity [5–9].
The newly developed PCR lateral flow method uses specific primers and capture probes for femA, mecA, and kdpC, respectively. A specific single-stranded biotinylated PCR product is produced by means of asymmetric PCR using biotinylated and non-biotinylated primers mixed at a fixed ratio. The capture probe, which binds specifically to the single-stranded biotinylated PCR product, is immobilized on a membrane and forms a complex there with the single-stranded biotinylated PCR product and colloidal gold-labeled streptavidin allowing PCR products to be evaluated visually.
The method allows rapid diagnosis in approximately 3 h and shows high sensitivity and specificity because of the PCR and capture probe. Furthermore, multiplex PCR allows amplification and detection of three genes at the same time. Furthermore, this method renders detection devices, such as the electrophoresis apparatus, transilluminators, and fluorescence detectors, unnecessary.
In the present study, we investigated the clinical usefulness of multiplex PCR lateral flow as a novel, rapid genetic testing method for direct MRSA detection from clinical samples.
MATERIALS AND METHODS
Materials
We used clinical specimens submitted by the Department of Pediatrics at Kitasato University Hospital over the period of December 2009 to February 2010. A total of 74 specimens were examined, the breakdown of which was as follows: 24 specimens in which MRSA was detected by bacteriological testing (14 sputum specimens, 3 heparinized blood specimens from PF (pediatric) and SN (anaerobic) blood culture bottles (Sysmex/bioMérieux) (blood specimens), 2 nasal swab specimens, 2 skin swab specimens, 1 stool specimen, 1 pus specimen, 1 leachate specimen), 12 specimens in which methicillin-susceptible S. aureus (MSSA) was detected (6 sputum specimens, 2 skin swab specimens, 2 pus specimens, 1 nasal swab specimen, 1 cerebrospinal fluid specimen), 22 specimens containing S. aureus (comment Staphylococcus sp.) that did not meet the criteria of our hospital for harvested bacteria (≥106 cfu/ml in sputum, ≥104 cfu/ml in urine) (14 sputum specimens, 5 throat swab specimens, 2 urine specimens, 1 oral specimen), and 16 specimens in which S. aureus was not detected (No S. aureus) (8 sputum specimens, 1 catheter blood specimen, 1 genitourinary catheter specimen, 2 throat swab specimens, 1 skin swab specimen, 1 bile specimen, 1 cerebrospinal fluid specimen, 1 stool specimen). After isolation culture, the clinical specimens were stored frozen at −20°C until analysis in an MR-CL38-PT refrigerator (Mitsubishi). The approval of the ethics committee of Kitasato University Hospital School of Medicine (B Ethics 09–30) was obtained.
Methods
Isolation and Culture
Isolation medium for routine testing of blood and anaerobic specimens (including catheters) used a Trypticase Soy Agar II with 5% Sheep blood (TSA II)/chocolate (CHO) agar medium, dollargalluskey-modified BTB (BTB) agar medium, and CDC agar medium (Becton Dickinson). TSA II and CHO agar medium and BTB agar medium were used as isolation media for upper and lower respiratory tract specimens. BTB agar medium, MacConkey II agar medium with sorbitol, DHL agar medium (Becton Dickinson), TCBS agar medium (Eiken), and modified Skirrow agar medium EX (Nissui) were used as isolation media for stool specimens. The culture method used involved streak separation using a platinum loop after application of the clinical specimens to the medium. The incubation conditions for media involved in S. aureus detection were as follows: aerobic, in an environment of 60% ± 5% humidity using an IS92 Incubator (Yamato) for TSA II and BTB agar medium; 5% CO2, in an environment of 85% ± 5% humidity and 35°C using a TE-HER CP series CO2 Incubator (Hirasawa) for CHO agar medium; colonies were identified after 24 h of incubation.
Identification of Species and Drug Susceptibility
A Pos Combo 3.1J Panel (Siemens) was used for analysis of the biochemical properties of S. aureus. Drug susceptibility, measured as the minimum inhibitory concentration (MIC), was determined using the MicroScan WalkAway-96 SI (SIEMENS), fully automatic bacteria system, which conforms to the microdilution method recommended by the Clinical and Laboratory Standards Institute [10, 11]. S. aureus was treated as MRSA if it displayed sensitivity to either oxacillin (MPIPC), CFX, or both antibiotics included in the Pos Combo 3.1J panel at concentrations: MPIPC/MIC ≥2 μg/ml and CFX/MIC ≥4 μg/ml.
Multiplex PCR Lateral Flow
MRSA SCCmec types in Japan have been reported to be mostly type II [12, 13]. The multiplex PCR lateral flow for MRSA was recently developed to allow simultaneous detection of three genes: femA to detect S. aureus, mecA to detect SCCmec, and kdpC to detect SCCmec type II [14].
Pretreatment of Clinical Specimens
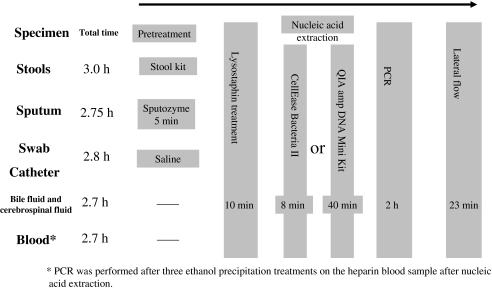
Figure 1 shows a schematic for PCR lateral flow operations. Stool specimens were treated with a QIAGEN Stool kit (Qiagen) and sputum samples were dispensed at one part purulent sputum to one part sputum lytic enzyme solution Sputozyme (Kyokuto) and homogenized. Swab and catheter specimens (throat swab, nasal swab, skin swab, pus, leachate, oral) were suspended in 700 μl of sterile saline. Along with the stock bile and cerebrospinal fluid, they were then treated with lysostaphin.
Fig. 1.
PCR lateral flow operation schematic. (Asterisk) PCR was performed after three ethanol precipitation treatments on the heparin blood sample after nucleic acid extraction.
Nucleic Acid Extraction
After completion of pretreatment, 6.6 μl of 10 ng/μl lysostaphin solution (Sigma) was added to 220 μl of clinical specimens and this was incubated at 37°C for 10 min [15, 16]. After completion of lysostaphin processing, nucleic acid was extracted from the clinical specimens using two nucleic acid extraction kits, the QIAamp DNA Mini Kit (Qiagen) and CellEaseII (Biocosm), and a comparison was conducted.
Nucleic Acid Extraction with the QIAamp DNA Mini Kit
After completion of processing with 200 μl of lysostaphin-treated sample, 50 μl of nucleic acid extract was produced from the clinical specimens using the QIAamp DNA Mini Kit (QIAGEN). The working time per specimen was approximately 40 min.
Nucleic Acid Extraction with the CellEaseII
Two microliters of CellEase A, 2 μl of CellEase B, and 5 μl of clinical sample pretreated with lysostaphin were added to a 2-ml tube; the mixture was heated at 68°C for 4 min and then at 94°C for 3 min using a thermal cycler and used as the nucleic acid extract. The working time per specimen was approximately 8 min.
Polymerase Chain Reaction
For polymerase chain reaction (PCR), 5 μl of nucleic acid extract and 25 μl of 2× Multiplex PCR Master Mix (Qiagen) reagent were added to a 200-μl tube, three primer sets adjusted to the concentrations shown below and distilled water were added and reacted with a total volume of 50 μl.
The primer set for detection of S. aureus used two forward primers (F), SauFemA-F (5′-ttacctatctctgctggtttcttcttt-3′) and SauFemA-F-3bio (5′-ttacctatctctgctggtttcttcttt-Biotin-3′), at a concentration ratio of 0.15 and 0.25 μM, respectively, and the reverse primer (R), SauFemA-R-bio (5′-Biotin-cactgcataacttccggcaaa-3′), was adjusted to a concentration of 0.70 μM.
The primer set for detection of SCCmec was similarly adjusted to concentrations of 0.15-μM MecA-F, 0.25-μM MecA-F-3bio (5′-ttagattgggatcatagcgtcattat-Biotin-3′), and 0.80-μM MecA-R-bio (5′-Biotin-aattccacattgtttcggtctaaaa-3′).
The primer set for detection of SCCmec type II was adjusted to concentrations of 0.15-μM KdpC-F (5′-cgcattgtcaaccgtaatatctg-3′), 0.25-μM KdpC-F-3bio (5′-cgcattgtcaaccgtaatatctg-Biotin-3′), and 0.50-μM KdpC-R-bio (5′-Biotin-caaactacggcaattcaaatcct-3′). These three primer sets were mixed in equal amounts and added to the tube as described above.
Thermal cycling conditions comprised 15 min at 95°C, followed by 50 cycles of 30 s at 94°C, 30 s at 60°C, 15 s at 72°C, followed by 2 min at 72°C. The PCR equipment used was a GeneAmp® PCR Systems 9700 (Applied Biosystems).
Detection
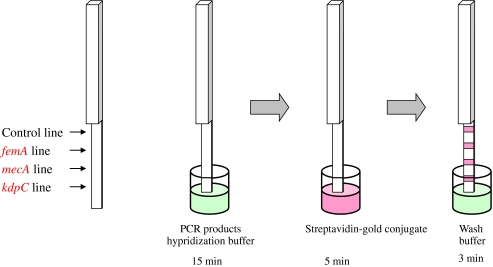
PCR products detected are shown in Fig. 2. Detection used lateral flow with a dipstick, which had a biotin-labeled control probe, the S. aureus detection probes SauFemA-1P (5′-ctggtggtacatcaaatgcattccgtca-3′) and SauFemA-2P (5′-ctggtggtacatcaaatgctttccgtca-3′), the SCCmec detection probe MecA-P (5′-ccaggaatgcagaaagaccaaagcataca-3′), and the SCCmec type II detection probe KdpC-P (5′-tctaaaccagagccagaagcggtcacc-3′) immobilized on the membrane. These probes immobilized on the membrane were performed as follows: these probes were spread on the membrane with brush, followed by membrane heat-treated at 80°C.
Fig. 2.
Lateral flow detection schematic.
Fourteen microliters of hybridization buffer and 20 μl of PCR product were added to each well of a 96-well plate and dipsticks left to soak for 15 min. Next, the dipstick was transferred to a second well to which streptavidin-gold conjugate (Funakoshi) had been added and left to stand for 5 min. Fifty microliters of wash buffer was then added and the dipstick was transferred to a third well. The dipstick was left to stand for 3 min and evaluation made from the red line obtained on the dipstick.
Evaluation
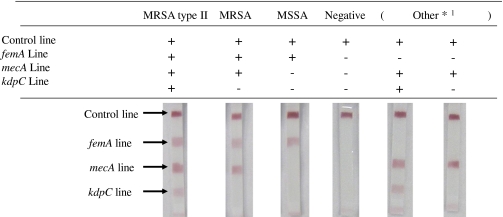
The evaluations of MRSA and other bacteria are shown in Table 1. Evaluation was made on the basis of the four red lines on the dipstick: the control line as a check, the femA line for S. aureus detection, the mecA line for SCCmec detection, and the kdpC line for SCCmec type II detection.
Table 1.
Evaluations of MRSA and Other Bacteria
*1 Other: non-S.aureus bacteria with SSCmec
S. intermedius, S. schleiferi, S. fleurettii, S. sciuri, S. epidermidis, S. capitis, S. hominis, S. vitulinus, S. pseudintermedius, S. saprophyticus, S. equorum, S. haemolyticus, S. saprophyticus, S. kloosii
Items for which only the control line was detected were classified as negative, those for which the control line and femA line were detected were classified as S. aureus, those for which the control line, femA line, and mecA line were detected were classified as MRSA, and those for which the control line, femA line, mecA line, and kdpC line were detected were classified as MRSA SCCmec type II. In addition, items for which the control line and mecA line or control line, mecA line, and kdpC line were detected were classified as non-S. aureus with SCCmec and were classified as other [17].
RESULTS AND DISCUSSION
Results
The results of PCR lateral flow and bacteriological testing are shown in Table 2. The results of analysis of the 74 clinical specimens from bacteriological testing and PCR lateral flow showed that among the 24 specimens identified as MRSA in bacteriological testing, 20 were identified as SCCmec type II MRSA, and two other types of MRSA by PCR lateral flow for a total of 22 specimens (91.7%), with two specimens negative for MRSA (8.3%). A blood specimen was deemed negative. Among the 12 specimens identified as MSSA from bacteriological testing (16.2%), PCR lateral flow showed one specimen to be SCCmec type II MRSA (8.3%) and 11 specimens to be MSSA (91.7%). In addition, among the 22 specimens identified as Staphylococcus sp. from bacteriological testing, PCR lateral flow identified six specimens as MRSA (27.3%) and showed ten specimens as MSSA (45.5%). Of the MRSA specimens, five were of SCCmec type II MRSA and one of other type MRSA. Among the 16 specimens identified as non-S. aureus from bacteriological testing, PCR lateral flow showed one specimen to be SCCmec type II MRSA (6.3%) and one specimen to be MSSA (6.3%). The results of the comparison of nucleic acids extracted using two nucleic acid extraction kits, the QIAamp DNA Mini Kit (Qiagen) and CellEaseII (Biocosm), showed differences between two of the clinical specimens identified as non-S. aureus from bacteriological testing. The two specimens with differences were extracted using the QIAamp DNA Mini Kit and classified as other (Table 2; others/non-S. aureus); extracts that used the CellEaseII showed negative results.
Table 2.
Results of PCR Lateral Flow and Bacteriological Testing
| N | MRSA | MSSA | Comments Staphylococcus sp. | No S. aureus | ||
|---|---|---|---|---|---|---|
| 24 | 12 | 22 | 16 | |||
| PCR lateral flow results | MRSA (SSCmec type II) | 91.7% (22) | 8.3% (1) | 27.3% (6) | 6.3% (1) | |
| (83.3% (20)) | (8.3% (1)) | (22.7% (5)) | (6.3% (1)) | |||
| MSSA | 0% (0) | 91.7% (11) | 45.5% (10) | 6.3% (1) | ||
| Others | 0% (0) | 0% (0) | 18.2% (4) | Qiagen | Biocosm | |
| 31.3% (5) | 18.8% (3) | |||||
| Negative | 8.3% (2) | 0% (0) | 9.1% (2) | Qiagen | Biocosm | |
| 56.3% (9) | 68.8% (11) | |||||
The test was repeated three times each sample; the results were always the same
DISCUSSION
In terms of speed, PCR is a far more useful test method than bacteriological testing for MRSA infections, which require early diagnosis and treatment [18]. However, running multiple PCRs in a single tube presents many difficulties from the point of view of sensitivity, specificity, and complexity. In the present study, we developed a novel multiplex PCR lateral flow method and investigated the clinical usefulness of the method in MRSA diagnosis. This method uses multiplex PCR to allow the PCR of three genes in the same specimen in a single tube. Furthermore, using the dipstick-type lateral flow method, the three genes can be detected simultaneously with a single stick. In order to create a single-stranded PCR product, F-3bio was added and single-stranded DNA was generated using complementarily associated F-3bio. In addition, sensitivity was improved in the presence of both R-bio and F-3bio. Single-stranded DNA was detected complementarily with a detection probe, thereby increasing specificity. As mentioned above, improvement in both sensitivity and specificity is a major feature of this method.
In general, PCR is subject to several problems that become major causes of contamination including the time for analysis, the operational processes, and the operational method for nucleic acid extraction [19]. In order to develop a more convenient extraction method with less contamination, we investigated the extraction of DNA from identical specimens and the detection of S. aureus with two widely used nucleic acid extraction kits, the QIAamp DNA Mini Kit (QIAGEN) and CellEaseII (Biocosm) [20]. The results showed no difference between detection results for the two kits for 72 of the 74 specimens. The remaining two specimens were identified as non-S. aureus by bacteriological testing and identified as “other” and “negative” by PCR using the different DNA extraction kits (Table 2). However, no difference was observed between the kits in discrimination of MRSA and MSSA. Furthermore, nucleic acid extraction time with CellEaseII was approximately 8 min in comparison to 40 min with QIAamp DNA Mini Kit, allowing the time to be reduced by approximately 32 min. This method was considered to allow extracts to be processed in an airtight state until addition to PCR, to have little risk of contamination, and to be a method well-suited to PCR lateral flow.
Three blood specimens identified as MRSA from bacteriological tests all initially showed as negative with PCR lateral flow. The PF (pediatric) blood culture bottle used for the blood specimens contained activated carbon particles, an antibiotic adsorbent agent. Since this inhibits the PCR, centrifugation was required [9]. In addition, three blood specimens came from the same patient and were taken during administration of 13,200 units/6 days of Novo Heparin (Mochida Pharmaceuticals) to treat disseminated intravascular coagulation. Blood specimens containing heparin are generally unsuitable for PCR [21] and the present results from these blood samples are believed to have been affected by heparin inhibition of PCR. We therefore carried out three ethanol precipitation treatments after nucleic acid extraction from these specimens and ran a PCR lateral flow. The results showed that MRSA was detected in two of the three specimens. Therefore, a nucleic acid extraction method to remove the influence of substances such as heparin that inhibit PCR and blood sampling using an EDTA salt as an anticoagulant may be necessary for detection from blood specimens.
In the present study, bacteriological testing identified S. aureus in 36 of the 74 specimens examined (48.6%), whereas PCR lateral flow identified S. aureus in 52 of the specimens (70.2%). Of 12 specimens identified as MSSA by bacteriological testing (16.2%), one was shown to be MRSA by PCR lateral flow (8.3%). Regarding the clinical specimens that did not meet the criteria of our hospital for harvested bacteria in bacteriological tests (≥106 cfu/ml in sputum, ≥104 cfu/ml in urine) and were identified as Staphylococcus sp. from report comments, six specimens were identified as MRSA (27.3%), and ten as MSSA (45.5%) by PCR lateral flow. Furthermore, among the clinical specimens identified as non-S. aureus from bacteriological testing, PCR lateral flow showed one specimen to be MRSA (6.3%) and one specimen to be MSSA (6.3%). Discrepancies in test results between the methods were believed to be attributable to differences in sensitivity. The MRSA and MSSA diagnostic concordance rate for PCR lateral flow and bacteriological testing was 91.7% overall and 95.2% when the PCR-inhibiting blood specimens are excluded. A discrepancy between bacteriological testing and PCR lateral flow which was observed for one nasal swab specimen and was believed to be due to a problem with the specimen’s treatment and storage. On the basis of our findings, PCR lateral flow was considered to have higher detection sensitivity than bacteriological testing. In the current situation of increasing MRSA infections, there is the risk that a carrier bearing quantities of bacteria difficult to detect by bacteriological testing may become an infection source for hospital-acquired infections. This will need attention in the control of hospital-acquired infection in the future, and we believe this method will be very useful as one of these controls.
Bacteria other than MRSA have also been reported to have the mecA and kdpC genes [22]. However, detection of the femA and mecA genes and mecA and kdpC genes was a prerequisite for identification of MRSA in the present study. It was believed, therefore, that bacteria other than MRSA had virtually no effect on test results from PCR lateral flow. In future, however, bacteriological testing and comparative studies should be carried out with more test materials and the effect of non-specific reactions due to bacteria other than MRSA should be investigated.
In conclusion, multiplex PCR lateral flow appears to be useful as a test method for MRSA detection with high sensitivity, specificity, and speed. It is feasible for clinical application in MRSA infections, which require early diagnosis and treatment.
Acknowledgments
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
REFERENCES
- 1.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 2.Center for Disease Control and Prevention (CDC) Four pediatric deaths from community-acquired meticillin-resistant Staphylococcus aureus -Minnesota and North Dakota, 1997–1999. Morbidity and Mortality; Weekly Report. 1999;48:707–710. [PubMed] [Google Scholar]
- 3.Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, Jamklang M, Chavalit T, Song JH, Hiramatsu K. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrobial Agents and Chemotherapy. 2006;50:1001–1012. doi: 10.1128/AAC.50.3.1001-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, Stobberingh EE. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clinical Microbiology and Infection. 2007;13:222–235. doi: 10.1111/j.1469-0691.2006.01573.x. [DOI] [PubMed] [Google Scholar]
- 5.Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clinical Infectious Diseases. 2008;46:787–794. doi: 10.1086/528716. [DOI] [PubMed] [Google Scholar]
- 6.Shibuya Y, Hara M, Higuchi W, Takano T, Iwao Y, Yamamoto T. Emergence of the community-acquired methicillin-resistant Staphylococcus aureus USA300 clone in Japan. Journal of Infection and Chemotherapy. 2008;14:439–441. doi: 10.1007/s10156-008-0640-1. [DOI] [PubMed] [Google Scholar]
- 7.Qian Q, Eichelberger K, Kirby JE. Rapid identification of Staphylococcus aureus in blood cultures by use of the direct tube coagulase test. Journal of Clinical Microbiology. 2007;45:2267–2269. doi: 10.1128/JCM.00369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higurashi Y, Okuzumi K. Detection methods for drug-resistant bacteria in routine examination—MRSA. Rinsho Byori. 2000;111:40–47. [PubMed] [Google Scholar]
- 9.Kimura T, Komori T, Hirose Y, Kurahashi S, Yamada Y, Kyotani N, Yuasa S, Fujita N. Evaluation of MRSA rapid detection by real-time PCR directly from positive blood cultures. Kansenshogaku Zasshi. 2010;84:199–205. doi: 10.11150/kansenshogakuzasshi.84.199. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Approved Standard eighth edition. CLSI document M7-A7. CLSI, Wayne, Pennsylvania.
- 11.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing. 15 (M100-S20). CLIS, Wayne, PA. [PubMed]
- 12.Ito T, Kuwahara K, Hisata K, Okuma K, Cui L, Hiramatsu K. Community-associated methicillin-resistant Staphylococcus aureus: current status and molecular epidemiological perspective. Kansenshogaku Zasshi. 2004;78:459–469. doi: 10.11150/kansenshogakuzasshi1970.78.459. [DOI] [PubMed] [Google Scholar]
- 13.Nada T, Yagi T, Ohkura T, Morishita Y, Baba H, Ohta M, Suzuki M. Usefulness of phage open-reading frame typing method in an epidemiological study of an outbreak of methicillin-resistant Staphylococcus aureus infections. Japanese Journal of Infectious Diseases. 2009;62:386–389. [PubMed] [Google Scholar]
- 14.Suzuki M, Tawada Y, Kato M, Hori H, Mamiya N, Hayashi Y, Nakano M, Fukushima R, Katai A, Tanaka T, Hata M, Matsumoto M, Takahashi M, Sakae K. Development of a rapid strain differentiation method for methicillin-resistant Staphylococcus aureus isolated in Japan by detecting phage-derived open-reading frames. Journal of Applied Microbiology. 2006;101:938–947. doi: 10.1111/j.1365-2672.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- 15.Yamazaki T, Endo K, Tominaga K, Fukuda M, Maesaki S, Hashikita G, Itabashi A. Epidemiological study of Arbekacin-resistant, methicillin resistant Staphylococcus aureus in Saitama Medical School Hospital. Kansenshogaku Zasshi. 2004;78:305–311. doi: 10.11150/kansenshogakuzasshi1970.78.305. [DOI] [PubMed] [Google Scholar]
- 16.Hirota Y, Okuzumi K, Shinji H. Low fibronectin-binding mutant of Staphylococcus aureus loses the ability to interact with host cells and tissues. Tokyo jikeikai Medical Journal. 2004;119:149–157. [Google Scholar]
- 17.Abraham JL, Morris DO, Griffeth GC, Shofer FS, Rankin SC. Surveillance of healthy cats and cats with inflammatory skin disease for colonization of the skin by methicillin-resistant coagulase-positive staphylococci and Staphylococcus schleiferi ssp. schleiferi. Veterinary Dermatology. 2007;18:252–259. doi: 10.1111/j.1365-3164.2007.00604.x. [DOI] [PubMed] [Google Scholar]
- 18.Ishiko H. Rapid identification of bacteria by PCR and hybridization. Nippon Rinsho. 1994;52:344–349. [PubMed] [Google Scholar]
- 19.Lo YM, Mehal WZ, Fleming KA. False-positive results and the polymerase chain reaction. Lancet. 1988;2:8612–8679. doi: 10.1016/s0140-6736(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 20.Jaffe RI, Lane JD, Albury SV, Niemeyer DM. Rapid extraction from and direct identification in clinical samples of methicillin-resistant staphylococci using the PCR. Journal of Clinical Microbiology. 2000;38:3407–3412. doi: 10.1128/jcm.38.9.3407-3412.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satsangi J, Jewell DP, Welsh K, Bunce M, Bell JI. Effect of heparin on polymerase chain reaction. Lancet. 1994;343:1509–1510. doi: 10.1016/S0140-6736(94)92622-0. [DOI] [PubMed] [Google Scholar]
- 22.Zhang K, Sparling J, Chow BL, Elsayed S, Hussain Z, Church DL, Gregson DB, Louie T, Louie JM. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. Journal of Clinical Microbiology. 2004;42:4947–4955. doi: 10.1128/JCM.42.11.4947-4955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]