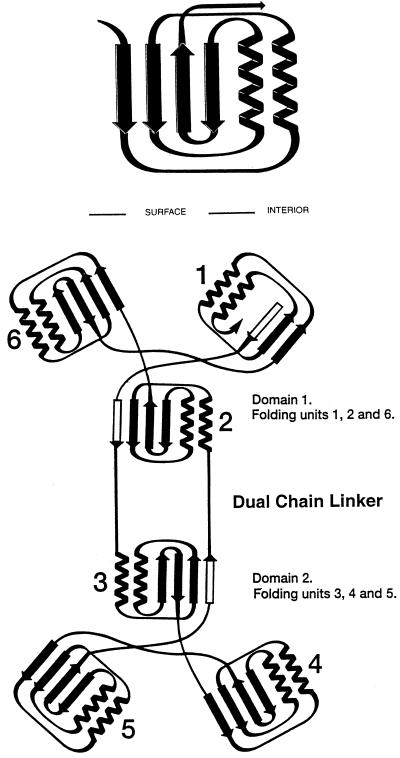
Figure 1.
Folding and topological symmetry of EPSPS (adapted from ref. 11). The two domain structure is formed by 6-fold replication of a protein folding unit (Upper) comprising two parallel helices and a four-stranded β-sheet. Each domain is formed from three of these folding units, which are related by an approximate 3-fold topological symmetry axis. In the open form of EPSPS, these axes are not collinear, but are presumably more so in the closed formed of the enzyme reported by Schönbrunn et al. (10). In each domain, three of the helices are buried and the surface of the molecule formed from the three β-sheets and the solvent-accessible faces of the other three helices. The N and C termini are located in Domain 1 with two crossover polypeptide segments creating a double hinge that links the two domains (Lower). Among the six folding units, three are folded from continuous segments of polypeptide chain. The other three contain the same arrangement of secondary structural features, but the sequences are not from a continuous chain. The arrangement positions the N termini of all 12 helices near the interface that results when the enzyme closes to form the active site. The positive macrodipoles from these helices presumably contribute to binding of substrates, products, and inhibitors, which are all multiply charged anions.