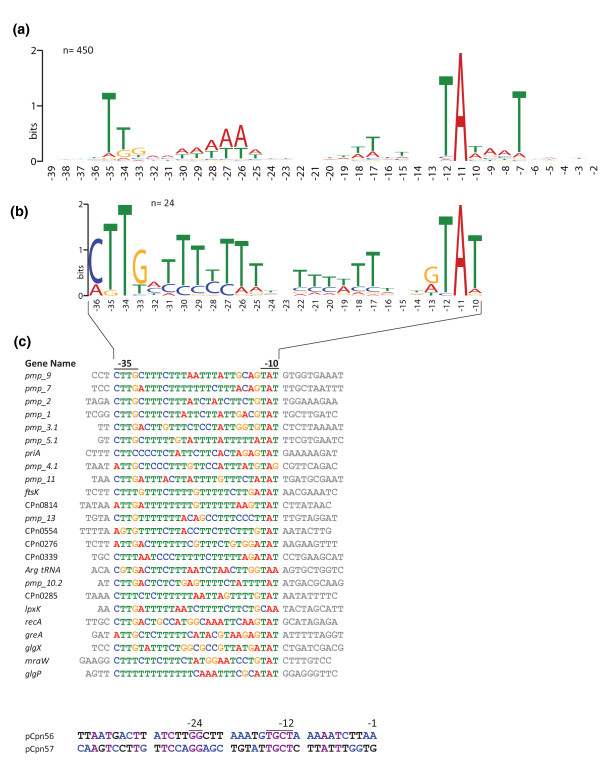
Figure 6.
Promoter motifs of C. pneumonia. Sequences upstream of 531 TSSs were extracted (positions -1 to -40) and common sequence motifs were searched using MEME software [45]. The coordinates give positions relative to TSSs. (a) The most prominent sequence alignment results from 450 out of 531 promoter sequences that were aligned. The motif resembles previously described σ66 promoter sequences and has the typical E. coli σ70 consensus sequences TATAAT at the -10 box. The -35 box is less conserved but is also similar to the E. coli consensus sequence TTGACA. For the promoter alignment a sequence shift of ±1 nucleotides from the TSS was permitted. (b) The second most prominent motif is derived from an alignment of 24 genes, of which 10 belong to the polymorphic outer membrane protein family (Pmp). The sequence contains well conserved -35 and -10 boxes and a T-rich spacer in between. (c) A search for the minimum conserved σ54 consensus sequence GG-N9-11-GC returned only two putative σ54 promoters that belong to sRNA candidates. No such sequence motif could be found in promoter regions of protein coding genes.