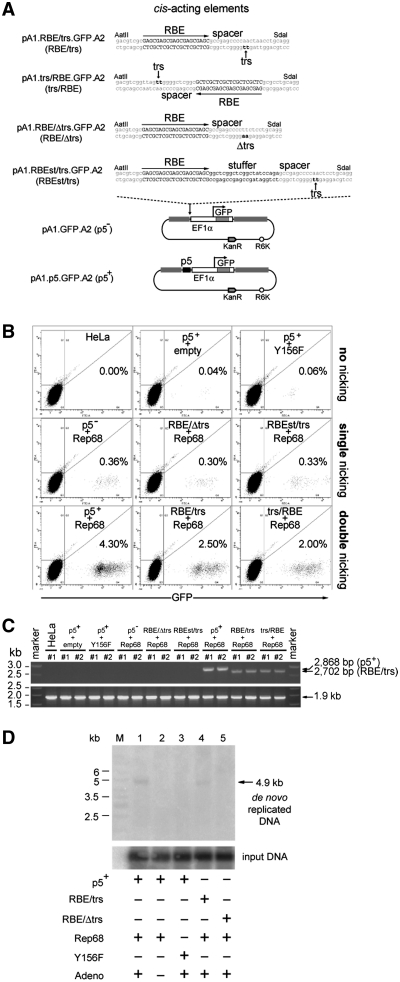
Figure 4.
Effect of DNA sequences susceptible or unsusceptible to AAV Rep-mediated nicking on the stable transfection levels with AAVS1-targeting vectors. (A) Structures of the nicking-competent targeting plasmids pA1.p5.GFP.A2, pA1.RBE/trs.GFP.A2 and pA1.trs/RBE.GFP.A2 and that of the nicking-resistant donor constructs pA1.GFP.A2, pA1.RBE/Δtrs.GFP.A2 and pA1.RBEst/trs.GFP.A2. The nucleotide sequence corresponding to the wild-type RBE is shown in black uppercase letters whereas the trs (i.e. the position at which Rep78/68-mediated nicking takes place) is indicated by black lowercase letters and a vertical arrow. The DNA sequence between the RBE and the trs is called the spacer. Targeting plasmid pA1.RBE/Δtrs.GFP.A2 has the dinucleotide at which Rep endonuclease-mediated nicking occurs mutated from TT to AA while in pA1.RBE.st/trs.GFP.A2 a 21-bp stuffer positions the trs at a bigger distance from RBE-bound AAV Rep molecules. For an explanation of the other symbols and elements see the legend of Figure 3A. (B) Representative dot plots of flow cytometric analysis of the frequency of GFP-modified HeLa cells at 37 days post-transfection in cultures initially exposed to pA1.p5.GFP.A2 and ‘empty’ plasmid (p5++empty), pA1.p5.GFP.A2 and pGAPDH.Rep68(Y156F) (p5++Y156F), pA1.GFP.A2 and pGAPDH.Rep68 (p5−+Rep68), pA1.RBE/Δtrs.GFP.A2 and pGAPDH.Rep68 (RBE/Δtrs+Rep68), pA1.RBEst/trs.GFP.A2 and pGAPDH.Rep68 (RBEst/trs+Rep68), pA1.p5.GFP.A2 and pGAPDH.Rep68 (p5++Rep68), pA1.RBE/trs.GFP.A2 and pGAPDH.Rep68 (RBE/trs+Rep68) or to pA1.trs/RBE.GFP.A2 and pGAPDH.Rep68 (trs/RBE+Rep68). Untransfected HeLa cells were used to set the background of the assay at 0.00% GFP-positive cells (HeLa). For each sample, 105 viable single cells were analyzed. (C) PCR analysis using primer set #649/#651 on chromosomal DNA extracted from untransfected HeLa cells (HeLa) and from HeLa cells co-transfected with pGAPDH.Rep68 (Rep68) and targeting constructs pA1.GFP.A2 (p5−), pA1.RBE/Δtrs.GFP.A2 (RBE/Δtrs), pA1.RBEst/trs.GFP.A2 (RBEst/trs), pA1.p5.GFP.A2 (p5+), pA1.RBE/trs.GFP.A2 (RBE/trs) or pA1.trs/RBE.GFP.A2 (trs/RBE). HeLa cells were also co-transfected with pA1.p5.GFP.A2 (p5+) and pGAPDH.Rep68(Y156F) or with an ‘empty’ control plasmid (empty). The genomic DNA was isolated at 43 days post-transfection. HPRT1-specific PCRs served as control for the integrity of the input DNA. (D) In vivo nicking assay based on Southern blot analysis of DpnI-resistant extrachromosomal DNA. Episomal DNA was isolated at 4 days post-transfection from 911 cells co-transfected with pGAPDH.Rep68 and pA1.p5.GFP.A2 (lanes 1 and 2), pGAPDH.Rep68 and pA1.RBE/trs.GFP.A2 (lane 4) or pGAPDH.Rep68 and pA1.RBE/Δtrs.GFP.A2 (lane 5). Episomal DNA isolated from 911 cells co-transfected with pGAPDH.Rep68(Y156F) and pA1.p5.GFP.A2 (lane 3) served as a negative control. All cell cultures except for the one represented by lane 2 were exposed to Ad.floxedΨ.F50 at a multiplicity of infection of 25 infectious units per cell. Lane M, Gene Ruler DNA Ladder Mix. Prior to Southern blot analysis, the DNA was digested with DpnI (to fragment the input prokaryotic DNA) and with NcoI. Southern blots were exposed to a radiolabeled GFP-specific probe. The position of the 4.9-kb GFP-containing NcoI fragments derived from de novo synthesized DNA is indicated by an arrow at the right of the autoradiogram.