Abstract
Mutations in mitochondrial DNA (mtDNA) are implicated in a broad range of human diseases and in aging. Compared to nuclear DNA, mtDNA is more highly exposed to oxidative damage due to its proximity to the respiratory chain and the lack of protection afforded by chromatin-associated proteins. While repair of oxidative damage to the bases in mtDNA through the base excision repair pathway has been well studied, the repair of oxidatively induced strand breaks in mtDNA has been less thoroughly examined. Polynucleotide kinase/phosphatase (PNKP) processes strand-break termini to render them chemically compatible for the subsequent action of DNA polymerases and ligases. Here, we demonstrate that functionally active full-length PNKP is present in mitochondria as well as nuclei. Downregulation of PNKP results in an accumulation of strand breaks in mtDNA of hydrogen peroxide-treated cells. Full restoration of repair of the H2O2-induced strand breaks in mitochondria requires both the kinase and phosphatase activities of PNKP. We also demonstrate that PNKP contains a mitochondrial-targeting signal close to the C-terminus of the protein. We further show that PNKP associates with the mitochondrial protein mitofilin. Interaction with mitofilin may serve to translocate PNKP into mitochondria.
INTRODUCTION
Single- and double-strand DNA breaks are induced directly by external and internal genotoxic agents such as ionizing radiation (IR), UV light and reactive oxygen species (ROS), or indirectly as a result of aborted topoisomerase action or during base excision repair (BER). Radiation and ROS-induced strand breaks frequently bear strand-break termini that require processing before ligation can occur, including 3′-phosphate and phosphoglycolate and 5′-hydroxyl end groups (1–3). Similarly, trapping of topoisomerase 1 by agents such as camptothecin or the presence of abasic sites or nicks adjacent to the cleavage site, followed by tyrosyl-DNA phosphodiesterase 1 (TDP1)-mediated cleavage of the covalent bond linking the DNA to the topoisomerase, generates single-strand breaks with 3′-phosphate and 5′-hydroxyl termini (4,5). BER performed by bifunctional DNA glycosylases, such as the Nei family members, NEIL1, NEIL2 and NEIL3, remove damaged bases and then cleave the DNA at the abasic sites through a lyase activity that involves β,δ-elimination to generate 3′-phosphate termini (6–8). The damaged DNA termini have to be restored to 3′-hydroxyl and 5′-phosphate functionality prior to the completion of the repair process by DNA polymerases and DNA ligases.
Polynucleotide kinase/phosphatase (PNKP) plays a major role in the restoration of correct DNA termini following strand cleavage by IR, ROS or NEIL-dependent BER (3,7,9,10). PNKP contains a forkhead-associated domain, which is a protein–protein interaction domain required for the association of PNKP with CK2-phosphorylated XRCC1 and XRCC4 (11–14), and independent DNA 3′-phosphatase and 5′-kinase domains (15,16). It has been shown that the DNA 3′-phosphatase activity of PNKP takes precedence over its DNA 5′-kinase activity in vitro (17). Downregulation of PNKP sensitizes cells to IR and hydrogen peroxide (18,19).
In addition to damage to nuclear DNA, mitochondrial DNA (mtDNA) is also subject to DNA damage. MtDNA is a 16.5 kbp circular molecule, encoding 37 genes including 13 proteins, 22 tRNAs and 2 rRNAs. Eukaryotic cells can have more than 100 mitochondria and each mitochondrion may contain 10 mtDNAs. In general, mtDNA constitutes about 1% of the total cellular DNA. ROS produced in relatively large quantities in mitochondria during respiration are the major source of mtDNA lesions (20). Damage to mtDNA, if not repaired, can develop into mutations, and mutations of the mtDNA are associated with different diseases including diabetes (21,22), cancer (23), neurodegenerative disorders (24) and aging (25). The rate of mutations in some regions of mtDNA is 20- to 100-fold higher than the nuclear DNA (26). This could be explained by the lack of protection of mtDNA by chromosomal proteins and the proximity of mtDNA to the inner membrane that contains the electron transport chain, which is a constant source of ROS (27).
As in the nucleus, BER is the main DNA repair pathway in mitochondria that deals with ROS-induced DNA lesions (8,27,28). Several DNA glycosylases have been identified in mitochondria including Nth and Nei family members (27). Mitochondria also contain a truncated form of APE-1 that can process abasic sites and DNA ends produced by β-elimination by DNA glycosylases/lyases such as NTH1 (29). In mitochondrial BER, replacement of missing nucleotides at damaged sites is performed by DNA polymerase γ (Polγ) instead of DNA polymerase β found in the nucleus (30), and ligation of DNA at single strand breaks is mediated by DNA ligase III (31). Recent studies have highlighted the importance of mtDNA ligase III for cell survival (31). Topoisomerase 1 and TDP1 are also present and functional in mitochondria (32). The presence of ROS, an active BER pathway and topoisomerase 1/TDP1 pathway point strongly to a need for PNKP or similarly acting protein to correct strand break termini in mitochondria.
Here, we demonstrate that functionally active full-length PNKP is present in mitochondria. Downregulation of PNKP results in a decrease of both mitochondrial and nuclear PNKP and accumulation of DNA damage in mtDNA, in addition to the previously documented increase in the nuclear DNA damage (18). Furthermore, we demonstrate that PNKP contains a C-terminal mitochondrial-targeting signal (MTS) (33,34). This C-terminal MTS is functional and is required for the localization of PNKP to mitochondria. Our results also indicate that PNKP associates with the mitochondrial protein mitofilin.
MATERIALS AND METHODS
Cell culture and transfection
A549 cells (human lung adenocarcinoma) and MCF7 cells (breast carcinoma) were obtained from ATCC (Manassas, VA, USA). PNKP-depleted cells were prepared by stable transfection of A549 cells with a pSuper expression vector containing an shRNA directed against PNKP as previously described (18). Cells were grown in DMEM/F12 nutrient mixture (1 : 1) and supplemented with 10% FBS, 50 U ml−1 penicillin, 50 µg ml−1 streptomycin, 2 mM l-glutamine, 0.1 mM non-essential amino acids and 1 mM sodium pyruvate (Invitrogen, Burlington, ON, USA) in a 5% CO2 humidified incubator at 37°C.
Cells were plated at ∼80–90% confluency in normal media without antibiotics and Lipofectamine 2000 was used the next day for transient transfections, according to the manufacturer's instructions (Invitrogen).
Immunoprecipitations and western blots
Cells were harvested from >90% confluent dishes by trypsinization, washed in cold PBS, pH 7.4. Cell pellets were resuspended in IP lysis buffer (200 mM NaCl, 2.5 mM MgCl2, 20 mM Tris–HCl pH 7.4 and 0.05% NP40) or lysis buffer (150 mM NaCl, 2 mM EDTA, 50 mM Tris–HCl, pH 7.4 and 1% NP40) plus 2 mM DTT, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (Sigma, St Louis, MO, USA), 1× complete protease inhibitor cocktail (Roche, Laval, QC, USA), followed by incubation on ice for 30–60 min. Next, cells were sonicated three times briefly in their corresponding buffer, centrifuged at 20 000g for 10 min at 4°C and the supernatants were snap-frozen and transferred to −80°C freezer for later use for either immunoprecipitation or western blots.
Cell lysates were precleared prior to immunoprecipitation by addition to Protein A-Sepharose beads (Sigma) and rotation at 4°C for 1 h followed by a spin to discard the beads. Then antibodies were added to the precleared lysates and rotated at 4°C for 1–3 h. Next, fresh beads were added to the lysate and rotation continued overnight. Finally, beads (now associated with antibodies) were harvested and washed three times with the lysis buffer, boiled in sample loading buffer and protein samples were run on a 10% PAGE gel. Samples were treated exactly the same for bead controls, except that no antibody was used. For immunoprecipitation of HA-tagged PNKP (HAPNKP), anti-HA affinity matrix (Roche) was used following the manufacturer’s recommendations. Western blots were performed using an ECL kit (Roche). Western blot images were scanned and quantified using ImageQuant for Windows version 5.2. The reading from the PNKP knock-down (KD) cell line in each experiment was normalized to the reading from A549 control cells. The P value was calculated using a two-tailed student's t test.
Antibodies used included polyclonal and monoclonal antibodies against PNKP (35), monoclonal mitofilin antibody (cat. # MSM02, Mitosciences, Eugene, OR), PCNA (cat. # 9857) and actin (cat. # SC1616, Santa Cruz Biotechnology, Santa Cruz, CA), HA (cat. # 12013819001, Roche, Brampton, ON), COX IV (ab16056-100) and VDAC1 (ab15895-100, Abcam, San Francisco, CA).
Generation of constructs and site-directed mutagenesis
RNAi-resistant PNKP was prepared by mutating PNKP cDNA at the shRNA targeting sequence, using the following primers:
Forward 5′-CAACCGGTTTCGAGAAATGACCGATTCCTCTC ATATCCCCG-3′
Reverse 5′-CGGGGATATGAGAGGAATCGGTCATTTC TCGAAACCGGTTG-3′.
The kinase negative PNKP was generated by site-directed mutagenesis using RNAi-resistant PNKP cDNA and the following primers:
Forward 5′-GGGATTCCCTGGGGCCGGGGCCTCCACCTTTCTCAAGAAGC-3′
Reverse 5′-GGGATTCCCTGGGGCCGGGGCCTCCACCTTTCTCAAGAAGC-3′
The phosphatase negative PNKP was generated by site-directed mutagenesis using RNAi-resistant PNKP cDNA and the following primers:
Forward 5′-AAGGTGGCTGGCTTTAATCTGAACGGGACGCTCATCACC-3′
Reverse 5′-GGTGATGAGCGTCCCGTTCAGATTAAAGCCAGCCACCTT-3′
The RNAi-resistant PNKP bearing a C-terminal haemagglutinin epitope tag (PNKPHA) was generated using RNAi-resistant PNKP cDNA and the following primers:
Forward 5′-TGACTGAATTCGCACCCAGGATGGGCGAGGTGGAGCCC-3′
Reverse 5′-TGACTGCGGCCGCTCAAGCGTAATCTGGAACATCGTATGGGTACGGGAGCCTCTTGACCGTC-3′.
The RNAi-resistant amino terminally-tagged HAPNKP was generated using RNAi-resistant PNKP cDNA and the following primers:
Forward 5′-TGACTGAATTCGCACCCAGGATGTACCCATACGATGTTCCAGATTACG CTGGCGAGGTGGAGCCCCCGG-3′
Reverse 5′-TGACT GCGGCCGCTCAGCCCTCGGA GAA CTG GC-3′.
The RNAi-resistant HAPNKP mutated at the mitochondrial-targeting signal (HAPNKP-mts) was generated by site-directed mutagenesis using cDNA from RNAi-resistant HAPNKP and the following primers:
Forward 5′-AACCCAGACGCCGCGAGCCGCGACGGGGACGTCCAGTGTGCCCGAGCC-3′
Reverse 5′- GGCTCGGGCACACTGGACGTCCCCGTCGCGGCTCGCGGCGTCTGGGTT-3′.
All of the above constructs were prepared in pIRESpuro3 from Clontech (Mountain View, CA, USA). Stable cell lines were prepared using the aforementioned cDNA constructs for transfection, followed by addition of 1 mM puromycin to the media and selection of surviving clones.
The constructs composed of the C-terminal region of PNKP, with and without the putative mitochondrial-targeting sequence, fused to GFP (CmtsPNKP + GFP and the CPNKP + GFP, respectively) were prepared using the pmEGFPN1 vector from Clontech. To prepare CmtsPNKP + GFP the following primers were used:
Forward 5′-TGACTAAGCTTGCACCCAGGATGGCCAGGTACGTCCAGTGTG-3′
Reverse 5′-GCTGATGGATCCCGGCCCTCGGAGAACTGGCAG-3′
To prepare CPNKP + GFP we used the reverse primer from CmtsPNKP + GFP and the forward primer 5′- TGACTAAGCTTGCACCCAGGATGGGCGTCCCCTGCCGCTG-3′
To mutate MTS in CmtsPNKP + GFP (mutCmtsPNKP + GFP) we used CmtsPNKP + GFP as a template. The site directed mutagenesis was accomplished using the following primers:
Forward 5′-GCTCAAGCTTGCACCCAGGATGGACGGGGACGTCCAGTGTGCCCGAGCC-3′
Reverse 5′-GGCTCGGGCACACTGGACGTCCCCGTCCATCCTGGGTGCAAGCTTGAGC-3′
We used QickChange II XL kit (cat. # 200521-5, Stratagene) for all site-directed mutagenesis experiments.
Purification of mitochondria, trypsin and proteinase K treatments
Mitochondria were purified essentially as described (36), with minor modifications. Briefly, cells grown in thirty 150-mm tissue culture dishes were harvested by trypsinization at >80% confluency, resuspended in hypotonic buffer (20 mM HEPES-KOH pH 7.4, 5 mM MgCl2, 5 mM KCl and 1 mM DTT) plus DTT and EDTA-free complete protease inhibitors (Roche) on ice (10 min), and homogenized in a Dounce homogenizer with 10–20 strokes. During homogenization cells were monitored by microscope to avoid break down of their nuclei. Immediately after homogenization, 2×MSH buffer (20 mM HEPES-KOH pH 7.4, 4 mM EDTA, 2 mM EGTA, 5 mM DTT, 420 mM mannitol, 140 mM sucrose) was added to the homogenate to stabilize the nuclei and samples were centrifuged two times at 1200g for 10 min each to prepare post-nuclear supernatant (PNS). The PNS then was centrifuged at 10 000 g to pull down crude mitochondria (CM). CM pellets were resuspended in 1×MSH/50% Percoll (Sigma) and loaded on top of a 1×MSH/50% Percoll column followed by ultracentrifugation at 50 000g for 70 min at 4°C. The white band of pure mitochondria that forms in the middle of the column was extracted by syringe, washed twice in 1×MSH and protease inhibitors, aliquoted, snap frozen and stored at −80°C. Prior to each experiment samples were thawed on ice and treated with trypsin (10 µg µl−1) for 20 min at room temperature followed by trypsin inhibitor treatment (29), or proteinase K (36) as required. An Artek Sonication Dismembrator model 150 was used to sonicate mitochondrial samples briefly when needed.
For isolation of rat liver mitochondria, normal Wistar rats were sacrificed and livers were removed quickly, rinsed with MSH buffer and diced. Small pieces of rat liver were homogenized by 10 strokes in a Dounce homogenizer and spun three times at 1200g for 10 min to remove cell debris. PNS was then treated the same as described for the cell lines.
Kinase and phosphatase assays of mitochondrial preparations
Kinase and phosphatase assays were performed essentially as described (18). Briefly, pure mitochondrial preparations were thawed on ice, trypsin-treated, pelleted by centrifugation, sonicated or not and resuspended in buffer C (70 mM sucrose, 210 mM mannitol, 80 mM succinic acid, pH 5.5, 10 mM MgCl2, 1 mM DTT and protease inhibitors) for kinase assays or in buffer D (70 mM sucrose, 210 mM mannitol, 50 mM Tris–HCl, pH 8.2, 10 mM MgCl2, 5 mM DTT, 1 mM spermidine) for phosphatase assays. A 21-mer oligonucleotide with a 5′-OH terminus and a 20-mer oligonucleotide bearing a 3′-phosphate group were used as the substrates for the kinase and phosphatase assays, respectively.
MtDNA repair assay using extra large-qPCR
H2O2 was added for 1 h to the media of PNKP knock down (KD) A549 cells transfected with different constructs or vector only (18). Whole-cell (genomic and mitochondrial) DNAs were extracted using a miniprep kit according to the manufacturer's instructions (Qiagen) from untreated (control) and H2O2-treated cells after 0, 0.5, 2 or 4 h of repair. Extra large qPCR (XL-qPCR) was performed using a Gene Amp kit (Applied Biosystems) following conditions and primers described (37). Fluorescence quantification of XL-qPCR products was achieved using Quant-iT Pico Green dsDNA assay kit (Invitrogen).
Immunofluorescence imaging
A549 and MCF7 cells were grown to 20% confluency on coverslips, fixed and stained as previously described (38). GFP constructs were transfected into A549 or MCF7 cells using Lipofectamine 2000 and cells were used for imaging within 24 h post-transfection. Fluorescently stained cells were viewed on a Zeiss LSM 710 laser scanning confocal microscope mounted on an AxioObserver Z1 inverted microscope (Carl Zeiss, Jena, Germany) with a plan Apochromat 40× (NA 1.3) oil immersion lens. Individual immunofluorescence channels were collected sequentially to avoid signal bleeding through. GFP and mitofilin (using an Alexa 488-conjugated secondary antibody) were detected using 488-nm laser excitation and collected with a bandpass filter of 493–552 nm. PNKP and COX IV were detected with a secondary antibody conjugated with rhodamine, which was imaged with 561 nm laser excitation and emission capture with a bandpass filter of 562–638 nm. DAPI-stained DNA was imaged with a 405 nm laser and a bandpass filter of 410–497 nm. Images were collected at Nyquist sample rate with pinhole of 1 Airy unit.
RESULTS
Full-length PNKP is present in mitochondria
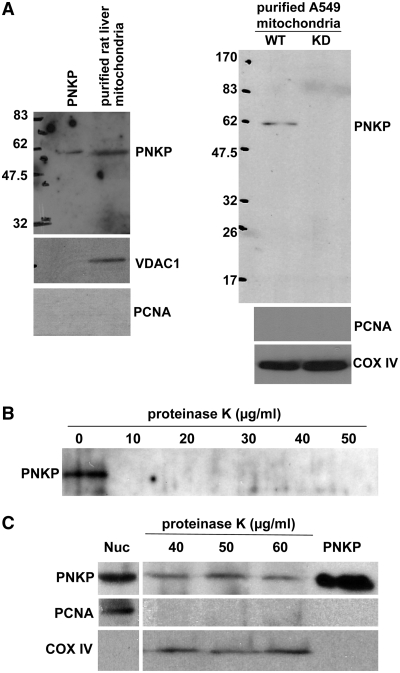
Mitochondria were purified from rat liver and human A549 lung cancer cells by centrifugation through a Percoll gradient (36), and then subjected to limited trypsin-treatment to improve the purity of the mitochondrial preparation by digesting extramitochondrial proteins (29). Western blot analysis of the mitochondrial preparation revealed the presence of apparently full-length PNKP (Figure 1A). The purity of the preparation was confirmed by the absence of PCNA as a nuclear marker and the presence of COX IV (cytochrome C oxidase—subunit IV) or VDAC1 (Voltage Dependent Anion Channel) as mitochondrial markers. Notably no PNKP was detectable in mitochondria isolated from A549 cells depleted of PNKP by shRNA. To further ensure that the PNKP signal came from inside the mitochondria and not due to contamination of the mitochondrial fraction with the nuclear PNKP, we also performed a proteinase K digestion assay. Purified human PNKP protein isolated from Escherichia coli was readily digested by proteinase K, being completely degraded by concentrations as low as 10 µg ml−1 (Figure 1B). However, mtPNKP recovered in the mitochondrial preparation was refractory to much higher concentrations of proteinase K (Figure 1C), indicating that the protein was protected by the mitochondrial membranes. Further titrations demonstrated that the mtPNKP was resistant to proteinase K digestion up to 400 µg ml−1 and significant digestion only started at 1 mgml−1 (data not shown). That the mtPNKP is full-length was also confirmed by purification of mitochondria from A549 cell lines stably expressing either N- or C-terminally HA-tagged PNKP followed by trypsin treatment (Supplementary Figure S1).
Figure 1.
Full-length PNKP is present in mitochondria. (A) Mitochondria were isolated from rat liver and wild-type (WT) and PNKP knock-down (KD) A549 cells. Mitochondrial protein extracts were immunoblotted with a monoclonal antibody to PNKP. Antibodies against PCNA (nuclear marker) and COX IV or VDAC1 (mitochondrial markers) were used to ensure the purity of the mitochondrial preparation. (B) Purified human PNKP protein is sensitive to proteinase K, and is completely digested even at the lowest concentration used (10 µg ml−1). (C) PNKP signal detected in purified mitochondria isolated from A549 cells is protected from proteinase K digestion. Proteins in nuclear extracts are shown on the left and purified PNKP protein on the right.
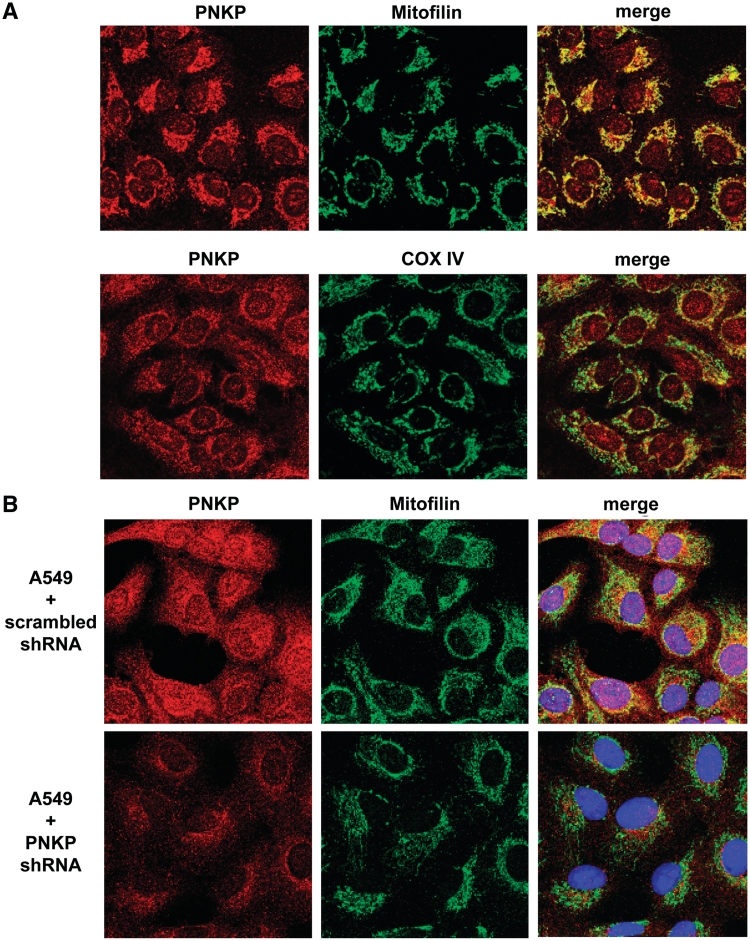
Next we employed immunofluorescence confocal microscopy to localize the endogenous PNKP in A549 cells (Figure 2). PNKP in both nuclei and mitochondria was detected with a polyclonal antibody to PNKP, and the mitochondrial PNKP signal colocalized with the mitochondrial markers mitofilin and COX IV (Figure 2A). Similar data were obtained with a monoclonal antibody to PNKP (Supplementary Figure S2). To further demonstrate the specificity of the signal for PNKP, we repeated the experiment with a PNKP KD A549 cell line (Figure 2B). Comparison of upper and lower panels in Figure 2B demonstrates a marked reduction of both the nuclear and mitochondrial signals in the PNKP KD cells.
Figure 2.
Endogenous PNKP colocalizes with mitochondrial markers. (A) Immunofluorescence of PNKP in A549 cells. PNKP localizes to mitochondria (in addition to the nuclei) as demonstrated by colocalization with the mitochondrial markers mitofilin and COX IV. (B) The shRNA KD of PNKP (lower panel) results in a decrease of the fluorescence signal for PNKP in both nuclei and mitochondria of A549 cells compared to the control cell line carrying scrambled shRNA (upper pannel).
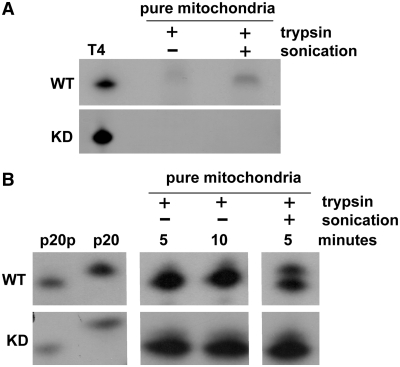
Based on western blotting mtPNKP appears to be the same size as the nuclear protein. However, we wanted to ensure that there is no major internal change in the functional domains of the enzyme that could interfere with their activities. We therefore directly examined the DNA kinase and phosphatase activities of mtPNKP (Figure 3A and B). Mitochondria were purified on a Percoll gradient, trypsin-treated and either sonicated or not before DNA kinase and phosphatase assays were performed (39). The kinase assay involves the transfer of radiolabeled phosphate from ATP to the 5′-terminus of an oligonucleotide, while the phosphatase assay monitors the loss of 3′-phosphate from a 5′-radiolabeled oligonucleotide (39). As shown in Figure 3, very minor kinase and phosphatase activities were observed in the absence of sonication of the mitochondrial membrane (probably due to release of mitochondrial proteins as a result of freeze-thawing of the samples), and both activities increased substantially following sonication. In contrast, no DNA kinase or phosphatase activity was detected with mitochondrial preparations isolated from PNKP KD cells (Figure 3). It is possible that other mitochondrial enzymes may have 5′-kinase or 3′-phosphatase activity (e.g. APE1), but presumably they do not act efficiently on the single-stranded substrates used to test PNKP activities. Thus this experiment indicates that mtPNKP possesses both DNA kinase and phosphatase activities and although we have not examined the protein at the amino acid sequence level, it would suggest that mtPNKP does not differ substantially, if at all, from the nuclear protein.
Figure 3.
Mitochondrial PNKP displays both DNA kinase and phosphatase activities. (A) DNA kinase activity was measured by transfer of 32P-phosphate from ATP to an oligonucleotide (21-mer) with a 5′-hydroxyl terminus. Mitochondria were purified from wild-type (WT) and PNKP KD A549 cells and treated with trypsin to digest any possible residual extramitochondrial protein contamination. Trypsin-treated mitochondria, either sonicated (+) or not sonicated (−), were used for DNA kinase assays. Sonication increased the DNA kinase activity of the purified mitochondrial preparation in WT cells. DNA kinase activity of T4 PNK is shown as a positive control. (B) DNA 3′-phosphatase activity in mitochondrial preparations from WT and KD A549 cells was determined by dephosphorylation of a 5′-32P-labelled 3′-phosphorylated oligonucleotide (p20p), as substrate, resulting in conversion of p20p to p20 (markers shown on left). Sonication of the purified mitochondrial preparation substantially increased DNA phosphatase activity in WT cells.
mtPNKP is required for mtDNA repair
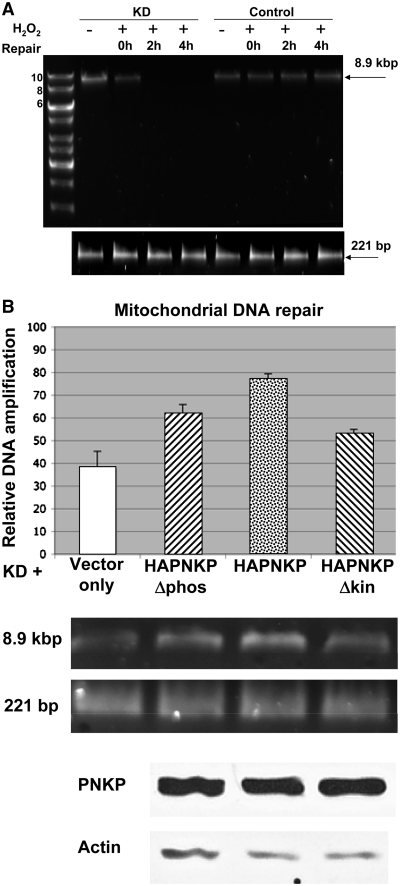
To show that mtPNKP is required for DNA repair in mitochondria, stable PNKP KD (A549 + PNKP shRNA) and control (A549 + scrambled shRNA) cell lines were exposed to 1.5 mM H2O2 for 1 h and repair was monitored 0, 2 and 4 h later. Total DNA was recovered from the cells (Supplementary Figure S3) and mtDNA repair was measured by (XL-qPCR) of an 8.9-kbp fragment of mtDNA (37). A 221-nt fragment of mtDNA was also amplified as an internal control and to show that the mtDNA was not degraded by H2O2, since such a small fragment would sustain only very limited damage under the conditions used. We employed picogreen to quantify the amount of large and small fragments amplified from mtDNA based on fluorescence reading. Finally, we compared the relative level of mtDNA amplification (large/small fragment) between the two cell lines. Our data (Figure 4A) clearly indicate that exposure of the PNKP-depleted (KD) cells to H2O2 ablated the signal from the 8.9-kbp fragment, strongly suggesting that PNKP is required for mtDNA repair. The decreased signal observed at 2 and 4 h in PNKP KD cell line (compared to the modest signal at 0 h) probably reflects an increase in the cleavage of mtDNA by DNA glycosylases/lyases during repair. Next, we wanted to determine the relative importance of the two enzymatic activities of PNKP for mtDNA repair in response to hydrogen peroxide. The PNKP KD cell line was transiently transfected with empty vector or HA-tagged wild-type PNKP (HAPNKP) as controls, and the kinase or phosphatase negative constructs (HAPNKPΔkin and HAPNKPΔphos), respectively. All the constructs carried mutations in the shRNA recognition site that prevented inhibition by shRNA but maintained the correct amino acid sequence (Supplementary Figure S4). The partially enzyme-inactivated PNKP constructs were prepared by site-directed mutagenesis of key residues in the kinase (K378A) or phosphatase (D171A and D173A) domains (17). The assay (Figure 4B) revealed intermediate activity for the kinase- and phosphatase-mutated PNKPs in dealing with H2O2-induced mtDNA damage in comparison to the wild-type PNKP and suggests that both the kinase and phosphatase activities of PNKP are required for the maintenance of mtDNA integrity.
Figure 4.
Functional PNKP is required for DNA repair in mitochondria following exposure to H2O2. (A) Total (i.e. nuclear + mitochondrial) DNA was purified from A549 cell lines stably transfected with PNKP shRNA (KD) or scrambled shRNA (control) (Supplementary Figure S3) following exposure to 1.5 mM H2O2 for 1 h and repair for 0, 2 and 4 h. Controls (−) were not exposed to H2O2. Upper panel: XL-qPCR performed on mtDNA, as described in Materials and Methods, amplified an 8.9-kbp PCR product following H2O2 exposure and repair in A549 cells expressing scrambled shRNA, but not in PNKP KD A549 cells. Lower panel: PCR of a 221-bp fragment from both cell lines indicating that both lines contain comparable amounts of mtDNA and that the H2O2 treatment did not degrade the DNA. (B) DNA repair in mitochondria in PNKP KD cells complemented with empty vector, wild-type (HAPNKP) or kinase (HAPNKPΔkin) or phosphatase inactive (HAPNKPΔphos) PNKP. XL-qPCR was used to examine the level of DNA repair 30 min after exposure to 1 mM H2O2 for 1 h. The signal from the amplified 8.9 kbp mtDNA following XL-qPCR was normalized to the signal from the 221-bp fragment using Quant-it Pico Green Assay Kit. Error bars show the SD for three independent experiments. The western blot shows that similar levels of PNKP protein were expressed in the PNKP KD background.
PNKP contains a functional mitochondrial-targeting signal close to its carboxy-terminus
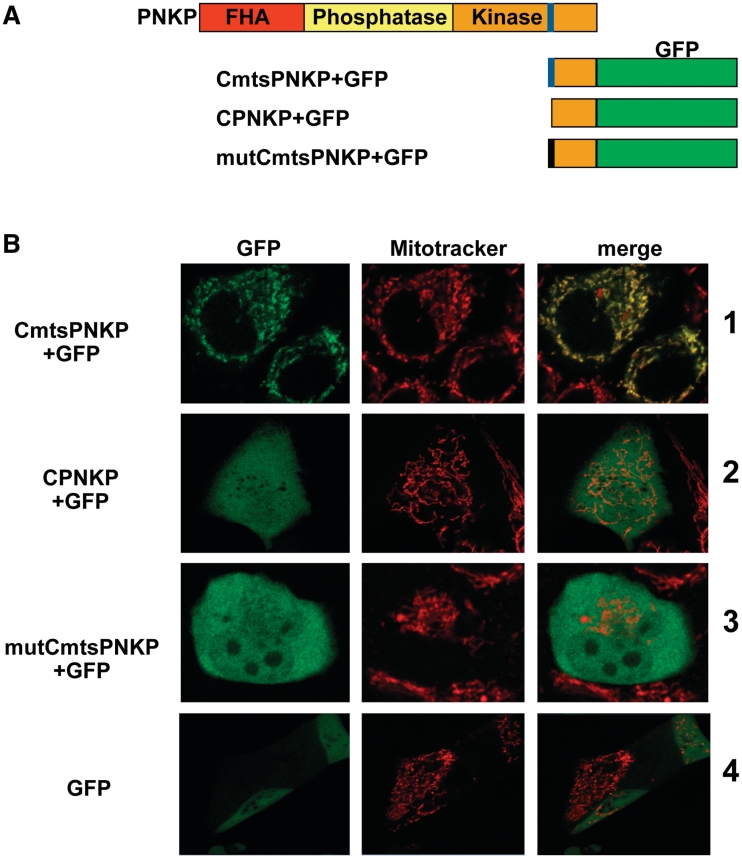
Mitochondrial-targeting signals (MTS) are most frequently found at the N-termini of proteins, but some of the mtDNA repair proteins reported to date do not contain an amino-terminal MTS. A computer-based analysis using multiple programs (including Mitoprot II and Predotar) showed that PNKP does not contain a canonical (N-terminal) MTS. However, a closer inspection of the sequence of PNKP revealed the presence of a ‘cryptic’ MTS close to the carboxy-terminus of the protein. This putative MTS consisted of amino acids 432–441 (ARYVQCARAA) and was identified by several computer programs, as mentioned. To determine if this MTS is functional, we fused the carboxy-terminus of PNKP with (CmtsPNKP) or without the MTS (CPNKP) to GFP, generating CmtsPNKP + GFP and CPNKP + GFP (Figure 5A). The constructs were then transfected into A549 (Figure 5B) and MCF7 (data not shown) cells. In both cases an additional methionine was added to the amino terminus of the fusion protein. The carboxy-terminal region of PNKP incorporating the MTS was functional as a mitochondrial-localization signal and transferred most of the expressed CmtsPNKP + GFP into the mitochondria (Figure 5B, row 1). However, when the MTS was not included with the carboxy-terminal region of PNKP no CPNKP + GFP was transferred into mitochondria (Figure 5B, row 2), a result similar to the situation seen with GFP alone (Figure 5B, row 4). Further confirmation was provided by mutating the first three amino acids of the MTS as follows: A432D, R433G and Y434D. Computer analysis indicated that these mutations would dramatically decrease the capacity of the identified MTS to function as a true mitochondrial-localization signal. We observed that the protein expressed by this mutated construct (mutCmtsPNKP + GFP) failed to localize to the mitochondria (Figure 5B, row 3).
Figure 5.
Mitochondrial localization of PNKP is dependent on the presence of a mitochondrial-targeting signal (MTS) in proximity to its carboxy terminus. (A) Computer programs (Mitoprot, Psort II and Predotar) predicted the presence of a mitochondrial-targeting signal (MTS) close to the C-terminus of PNKP (shown in blue). To further examine this potential MTS we generated three constructs (i) CmtsPNKP + GFP containing the GFP fused to the PNKP C-terminus including the putative MTS, (ii) CPNKP + GFP, which is essentially the same as CmtsPNKP + GFP but lacking the MTS sequence and (iii) mutCmtsPNKP + GFP, a mutated form of CmtsPNKP + GFP with the first three amino acids of the putative PNKP MTS mutated as follows: A432D, R433G and Y434D. In all cases a methionine was included at the amino-terminus. (B) The constructs were transfected into A549 cells and the cellular localization of GFP was monitored. Only the GFP fusion protein containing the wild-type MTS localized to mitochondria (row 1) as shown by colocalization with Mitotracker Orange (Molecular Probes). CPNKP + GFP, mutCmtsPNKP + GFP and GFP alone showed a diffuse signal throughout the cell (rows 2–4).
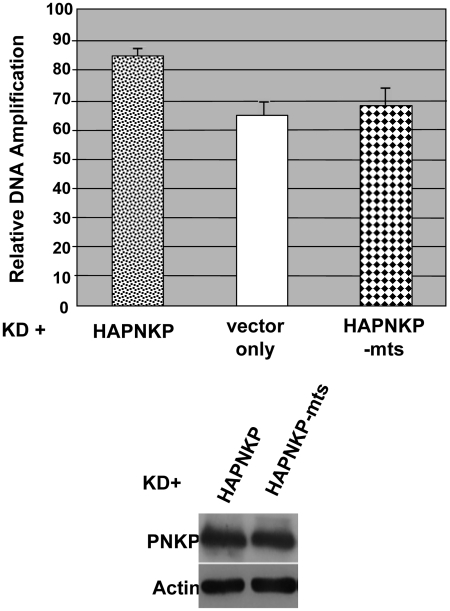
To test if the MTS is functional in the context of full-length PNKP, XL-qPCR was used to compare the functionality of HAPNKP-mts (MTS-mutated form of HAPNKP incorporating the same mutations to the MTS as described above) to HAPNKP in mtDNA repair. HAPNKP (positive control), vector only and HAPNKP-mts were transfected into PNKP KD cell lines prepared from A549 cells. As shown in Figure 6, the loss of the MTS resulted in a clear decrease in the activity of PNKP during mtDNA repair, similar to transfection with the vector only. The control western blot showed that the level of expression of HAPNKP-mts was the same as HAPNKP and thus the observed effect was not due to a lower level of protein expression of the mutated form of PNKP.
Figure 6.
The MTS of PNKP is required for its function in mtDNA repair. XL-qPCR was used to monitor mtDNA repair in PNKP-depleted A549 cells treated with hydrogen peroxide as described in Figure 4B. Transient complementation of the cells with PNKP mutated in the first three amino acids of the MTS (HAPNKP-mts), as opposed to the wild-type protein (HAPNKP), reduces DNA repair in mitochondria to a level similar to the vector only control. The western blot of whole-cell extracts shown at the bottom of the figure indicates that similar levels of HAPNKP-mts and HAPNKP proteins were expressed in the A549 cells.
PNKP interacts with the mitochondrial proteins
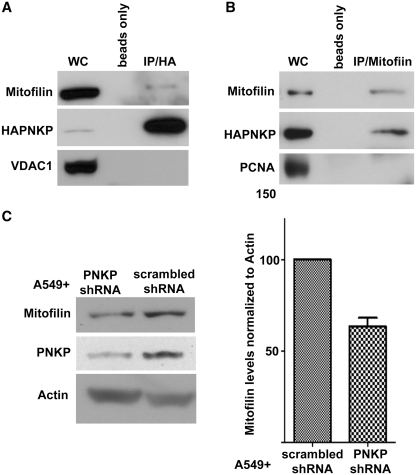
It would be expected that the presence of PNKP in mitochondria would give rise to interaction of mtPNKP with mitochondrial proteins. We therefore expressed HAPNKP in an A549 PNKP KD background and immunoprecipitated the PNKP using anti-HA antibodies. We were able to identify mitofilin as one of the mitochondrial proteins that coimmunopreciptated with PNKP (Figure 7A). Interaction with mitofilin was further confirmed by reciprocal immunoprecipitation of HAPNKP by antibodies to mitofilin (Figure 7B). Mitofilin is a transmembrane protein of the inner membrane of mitochondria that has been shown to interact with PARP-1 (40), and DISC1 (disrupted-in-schizophrenia 1) (41). Interestingly, western blot analysis indicated that downregulation of PNKP by 70–90% in A549 cells reduced the level of mitofilin (Figure 7C). A similar reduction of the level of mitofilin in DISC1 KD cells has also been reported (41).
Figure 7.
PNKP physically interacts with mitochondrial proteins. (A) Coimmunopreciptation of mitofilin by anti-HA antibody from whole-cell extract (WC) of A549 cells expressing HAPNKP. An antibody to the mitochondrial protein VDAC1 was used to ensure no non-specific pull-down of mitochondrial proteins. (B) Coimmunoprecipitation of PNKP with mitofilin from whole-cell extract of A549 cells expressing HAPNKP using a monoclonal antibody to mitofilin to immunoprecipitate mitofilin. In this case, the immunopreciptate was also tested for the presence of PCNA as a marker of potential contamination by nuclear proteins. The control lane ‘beads only’ indicates that no PNKP bound to the Sepharose beads in the absence of antibodies. (C) KD of PNKP by shRNA results in a decrease of about 40% in cellular mitofilin content. The graph shows the mean and SD values from three independent determinations (P = 0.013).
DISCUSSION
PNKP is a bifunctional end-processing enzyme involved in BER, and repair of single- and double-strand breaks. PNKP has been reported to be a nuclear protein by us and other laboratories (39,42). Here, we provide evidence that PNKP is also found in mitochondria. Given the abundance of ROS and the presence of other BER and single strand break repair proteins in mitochondria including uracil-DNA glycosylase, NEIL1, TDP1, APE1 and DNA ligase III (27,28,32), the appearance of PNKP in mitochondria is perhaps not surprising. Our data indicate that mtPNKP is the same size as the nuclear protein. Several of the BER proteins found in the mitochondria differ from their nuclear isoforms due to alternative splicing, as seen with mitochondrial uracil-DNA glycosylase (43), or post-translational processing, as observed with mitochondrial APE1 (29), or through the use of an alternative translation-initiation start site, as seen with mtDNA ligase III (44). Based on the western blots (Figure 1) coupled with observing both N- and C-terminally HA-tagged PNKP in the mitochondria (Supplementary Figure S1), we conclude that mtPNKP appears to be the same protein as nuclear PNKP.
Mitochondrial proteins often differ from their nuclear counterparts to incorporate a mitochondrial-targeting sequence (MTS) or to shed a nuclear localization signal (NLS). About 50% of mitochondrial proteins contain an MTS at their amino terminus. For example, aprataxin, which has recently been shown to play a role in mtDNA repair, possesses an amino-terminal MTS (45). However, localization to mitochondria does not necessarily require an amino-terminal MTS (46). Several mtDNA repair proteins, such as TDP1 and PARP-1, appear to lack an MTS (32,40). An MTS has been identified at the C-terminus of APE1 (47), and it appears that the redirection of APE1 into the mitochondria occurs after proteolytic removal by a mitochondria-associated N-terminal peptidase of the first 33 amino acids of APE1 that contains an NLS (29). A putative NLS has been previously identified in PNKP (amino acids 301–304), but it lies in the phosphatase domain of the protein (39,42,48,49) and is presumably retained in the mitochondrial PNKP. Alternatively, it has been shown that a C-terminal MTS can direct proteins to mitochondria in a carboxy to amino-terminal direction (34,50), and C-terminal MTSs have been found in several other mtDNA processing enzymes including the yeast DNA helicase Hmi1p (33) and the human nuclease/helicase DNA2 (51). An in silico analysis of the PNKP sequence uncovered a putative MTS located near the carboxy-terminus of the protein. We determined that this sequence is a functional MTS required for PNKP repair activity in mitochondria (Figure 6). It was also able to target GFP to mitochondria when placed at the N-terminus of GFP (Figure 5). Interestingly, using a similar in silico analysis with Mitoprot II and Predotar software, we detected a potential MTS in PARP-1 (amino acids 846–860) but this will require experimental verification.
We observed that mtPNKP retains both the DNA kinase and phosphatase activities (Figure 3), and that both activities are required for timely repair of mtDNA following damage induced by hydrogen peroxide (Figure 4). We have previously shown that shRNA-mediated depletion of PNKP sensitizes A549 cells to hydrogen peroxide (18). Further experimentation will be required to determine the relative importance of nuclear versus mitochondrial PNKP in response to hydrogen peroxide and other ROS, since this may have implications for the recently identified autosomal recessive neurological disorder, MCSZ, associated with mutations in PNKP (52). Although a direct comparison of the relative importance of PNKP kinase versus phosphatase activity for the repair of nuclear DNA has yet to be carried out, the phosphatase activity has been shown to play a critical role in single-strand break repair following oxidative damage by hydrogen peroxide and IR (19,53). The phosphatase activity of PNKP is also significantly more active than the kinase activity (17). Oxidative damage is more frequently associated with modifications to DNA 3′-strand-break termini than the 5′-termini (1,2). That the expression of a kinase-mutated PNKP had a similar outcome as the phosphatase-mutated PNKP on the repair of mtDNA suggests that 5′-hydroxyl termini are generated in similar quantities to modified 3′-termini, maybe through the intervention of trapped topoisomerase I complexes, or it may reflect differing levels of other mitochondrial repair proteins that may compensate for reduced PNKP activity. Alternatively, the relative importance of the kinase and phosphatase activities might be skewed by incomplete shRNA-mediated downregulation of endogenous PNKP that leaves cells with sufficient levels of the DNA phosphatase activity to partially compensate for the loss of phosphatase activity when expressing the phosphatase-mutated PNKP.
Our examination of an interaction between PNKP and mitofilin was predicated on a recent finding by Rossi et al. (40) that mitochondrial PARP-1 interacts with mitofilin. These authors further established that the presence of PARP-1 in mitochondria was dependent on the level of mitofilin and, based on the observation that mitofilin associates with proteins implicated in protein import into the mitochondria (54), they suggested that mitofilin may be involved in the translocation of PARP-1 across the mitochondrial membranes. Thus, interaction of mitofilin with PNKP could also assist with mitochondrial localization of PNKP. Knock down of PNKP resulted in a statistically significant decrease in the level of mitofilin. It is not readily apparent why the absence of PNKP should affect the level of mitofilin, but similar observations have been made with two other proteins, DISC1 (41) and the inner mitochondrial membrane protein ChChd3 (55). In the case of DISC1, its downregulation appears to increase proteasomal-mediated proteolysis of mitofilin (41). It remains to be determined whether downregulation of PNKP affects the level of mitofilin at the transcriptional or post-transcriptional level.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
Canadian Institutes of Health Research (grant number 15385 to M.W.); a postdoctoral fellowship from the Alberta Cancer Foundation and an Incentive award from the Alberta Heritage Foundation for Medical Research (to N.T.). Funding for open access charge: Canadian Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Xuejun Sun and Geraldine Barron for assistance with cell imaging, Dr Aghdass Rasouli-Nia, Mesfin Fanta, Ismail Abdou and Todd Mereniuk for other technical assistance and Dr Feridoun Karimi-Busheri for useful discussion and advice.
REFERENCES
- 1.Henner WD, Grunberg SM, Haseltine WA. Sites and structure of gamma radiation-induced DNA strand breaks. J. Biol. Chem. 1982;257:11750–11754. [PubMed] [Google Scholar]
- 2.Lennartz M, Coquerelle T, Bopp A, Hagen U. Oxygen—effect on strand breaks and specific end-groups in DNA of irradiated thymocytes. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1975;27:577–587. doi: 10.1080/09553007514550611. [DOI] [PubMed] [Google Scholar]
- 3.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem. Sci. 2011;36:262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pourquier P, Pilon AA, Kohlhagen G, Mazumder A, Sharma A, Pommier Y. Trapping of mammalian topoisomerase I and recombinations induced by damaged DNA containing nicks or gaps. Importance of DNA end phosphorylation and camptothecin effects. J. Biol. Chem. 1997;272:26441–26447. doi: 10.1074/jbc.272.42.26441. [DOI] [PubMed] [Google Scholar]
- 5.Plo I, Liao ZY, Barcelo JM, Kohlhagen G, Caldecott KW, Weinfeld M, Pommier Y. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair. 2003;2:1087–1100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 6.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair. 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 7.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M, Tomkinson AE, et al. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: evidence for a repair complex in human cells. DNA Repair. 2006;5:1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allinson SL. DNA end-processing enzyme polynucleotide kinase as a potential target in the treatment of cancer. Future Oncol. 2010;6:1031–1042. doi: 10.2217/fon.10.40. [DOI] [PubMed] [Google Scholar]
- 11.Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, Sarno S, Meggio F, Pinna LA, Caldecott KW. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell. 2004;117:17–28. doi: 10.1016/s0092-8674(04)00206-5. [DOI] [PubMed] [Google Scholar]
- 12.Koch CA, Agyei R, Galicia S, Metalnikov P, O'Donnell P, Starostine A, Weinfeld M, Durocher D. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu M, Mani RS, Karimi-Busheri F, Fanta M, Wang H, Litchfeld DW, Weinfeld M. Independent mechanisms of stimulation of polynucleotide kinase/phosphatase by phosphorylated and non-phosphorylated XRCC1. Nucleic Acids Res. 2010;38:510–521. doi: 10.1093/nar/gkp1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mani RS, Yu Y, Fang S, Lu M, Fanta M, Zolner AE, Tahbaz N, Ramsden DA, Litchfield DW, Lees-Miller SP, et al. Dual modes of interaction between XRCC4 and polynucleotide kinase/phosphatase: implications for nonhomologous end joining. J. Biol. Chem. 2010;285:37619–37629. doi: 10.1074/jbc.M109.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pheiffer BH, Zimmerman SB. 3′-phosphatase activity of the DNA kinase from rat liver. Biochem. Biophys. Res. Commun. 1982;109:1297–1302. doi: 10.1016/0006-291x(82)91918-0. [DOI] [PubMed] [Google Scholar]
- 16.Habraken Y, Verly WG. Further purification and characterization of the DNA 3′-phosphatase from rat-liver chromatin which is also a polynucleotide 5′-hydroxyl kinase. Eur. J. Biochem. 1988;171:59–66. doi: 10.1111/j.1432-1033.1988.tb13758.x. [DOI] [PubMed] [Google Scholar]
- 17.Dobson CJ, Allinson SL. The phosphatase activity of mammalian polynucleotide kinase takes precedence over its kinase activity in repair of single strand breaks. Nucleic Acids Res. 2006;34:2230–2237. doi: 10.1093/nar/gkl275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasouli-Nia A, Karimi-Busheri F, Weinfeld M. Stable down-regulation of human polynucleotide kinase enhances spontaneous mutation frequency and sensitizes cells to genotoxic agents. Proc. Natl Acad. Sci. USA. 2004;101:6905–6910. doi: 10.1073/pnas.0400099101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breslin C, Caldecott KW. DNA 3′-phosphatase activity is critical for rapid global rates of single-strand break repair following oxidative stress. Mol. Cell Biol. 2009;29:4653–4662. doi: 10.1128/MCB.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ralph SJ, Rodriguez-Enriquez S, Neuzil J, Saavedra E, Moreno-Sanchez R. The causes of cancer revisited: “mitochondrial malignancy” and ROS-induced oncogenic transformation—why mitochondria are targets for cancer therapy. Mol. Aspects Med. 2010;31:145–170. doi: 10.1016/j.mam.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Akbari M, Skjelbred C, Folling I, Sagen J, Krokan HE. A gel electrophoresis method for detection of mitochondrial DNA mutation (3243 tRNA(Leu (UUR))) applied to a Norwegian family with diabetes mellitus and hearing loss. Scand. J. Clin. Lab. Invest. 2004;64:86–92. doi: 10.1080/00365510410004209. [DOI] [PubMed] [Google Scholar]
- 22.van den Ouweland JM, Lemkes HH, Ruitenbeek W, Sandkuijl LA, de Vijlder MF, Struyvenberg PA, van de Kamp JJ, Maassen JA. Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat. Genet. 1992;1:368–371. doi: 10.1038/ng0892-368. [DOI] [PubMed] [Google Scholar]
- 23.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 24.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 26.Pesole G, Gissi C, De Chirico A, Saccone C. Nucleotide substitution rate of mammalian mitochondrial genomes. J. Mol. Evol. 1999;48:427–434. doi: 10.1007/pl00006487. [DOI] [PubMed] [Google Scholar]
- 27.de Souza-Pinto NC, Wilson DM, III, Stevnsner TV, Bohr VA. Mitochondrial DNA, base excision repair and neurodegeneration. DNA Repair. 2008;7:1098–1109. doi: 10.1016/j.dnarep.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu P, Demple B. DNA repair in mammalian mitochondria: much more than we thought? Environ. Mol. Mutagen. 2010;51:417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- 29.Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, Mitra S. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 2006;34:2067–2076. doi: 10.1093/nar/gkl177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen AB, Griner NB, Anderson JP, Kujoth GC, Prolla TA, Loeb LA, Glick E. Mitochondrial DNA integrity is not dependent on DNA polymerase-beta activity. DNA Repair. 2006;5:71–79. doi: 10.1016/j.dnarep.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, Van Houten B, Shuman S, McKinnon PJ, Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–248. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das BB, Dexheimer TS, Maddali K, Pommier Y. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc. Natl Acad. Sci. USA. 2010;107:19790–19795. doi: 10.1073/pnas.1009814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee CM, Sedman J, Neupert W, Stuart RA. The DNA helicase, Hmi1p, is transported into mitochondria by a C-terminal cleavable targeting signal. J. Biol. Chem. 1999;274:20937–20942. doi: 10.1074/jbc.274.30.20937. [DOI] [PubMed] [Google Scholar]
- 34.Folsch H, Gaume B, Brunner M, Neupert W, Stuart RA. C- to N-terminal translocation of preproteins into mitochondria. EMBO J. 1998;17:6508–6515. doi: 10.1093/emboj/17.22.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fanta M, Zhang H, Bernstein N, Glover M, Karimi-Busheri F, Weinfeld M. Production, characterization, and epitope mapping of monoclonal antibodies against human polydeoxyribonucleotide kinase. Hybridoma. 2001;20:237–242. doi: 10.1089/027245701753179811. [DOI] [PubMed] [Google Scholar]
- 36.Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair. 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Santos JH, Meyer JN, Mandavilli BS, Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 38.Crampton N, Kodiha M, Shrivastava S, Umar R, Stochaj U. Oxidative stress inhibits nuclear protein export by multiple mechanisms that target FG nucleoporins and Crm1. Mol. Biol. Cell. 2009;20:5106–5116. doi: 10.1091/mbc.E09-05-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karimi-Busheri F, Daly G, Robins P, Canas B, Pappin DJ, Sgouros J, Miller GG, Fakhrai H, Davis EM, Le Beau MM, et al. Molecular characterization of a human DNA kinase. J. Biol. Chem. 1999;274:24187–24194. doi: 10.1074/jbc.274.34.24187. [DOI] [PubMed] [Google Scholar]
- 40.Rossi MN, Carbone M, Mostocotto C, Mancone C, Tripodi M, Maione R, Amati P. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J. Biol. Chem. 2009;284:31616–31624. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park YU, Jeong J, Lee H, Mun JY, Kim JH, Lee JS, Nguyen MD, Han SS, Suh PG, Park SK. Disrupted-in-schizophrenia 1 (DISC1) plays essential roles in mitochondria in collaboration with Mitofilin. Proc. Natl Acad. Sci. USA. 2010;107:17785–17790. doi: 10.1073/pnas.1004361107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jilani A, Ramotar D, Slack C, Ong C, Yang XM, Scherer SW, Lasko DD. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem. 1999;274:24176–24186. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 43.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sykora P, Croteau DL, Bohr VA, Wilson DM., III Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc. Natl Acad. Sci. USA. 2011;108:7437–7442. doi: 10.1073/pnas.1100084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker MJ, Frazier AE, Gulbis JM, Ryan MT. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 2007;17:456–464. doi: 10.1016/j.tcb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 47.Li M, Zhong Z, Zhu J, Xiang D, Dai N, Cao X, Qing Y, Yang Z, Xie J, Li Z, et al. Identification and characterization of mitochondrial targeting sequence of human apurinic/apyrimidinic endonuclease 1. J. Biol. Chem. 2010;285:14871–14881. doi: 10.1074/jbc.M109.069591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galicia S, Koch CA, Cass CE, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 49.Meijer M, Karimi-Busheri F, Huang TY, Weinfeld M, Young D. Pnk1, a DNA kinase/phosphatase required for normal response to DNA damage by gamma-radiation or camptothecin in Schizosaccharomyces pombe. J. Biol. Chem. 2002;277:4050–4055. doi: 10.1074/jbc.M109383200. [DOI] [PubMed] [Google Scholar]
- 50.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, et al. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen J, Gilmore EC, Marshall CA, Haddadin M, Reynolds JJ, Eyaid W, Bodell A, Barry B, Gleason D, Allen K, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat. Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freschauf GK, Karimi-Busheri F, Ulaczyk-Lesanko A, Mereniuk TR, Ahrens A, Koshy JM, Rasouli-Nia A, Pasarj P, Holmes CF, Rininsland F, et al. Identification of a small molecule inhibitor of the human DNA repair enzyme polynucleotide kinase/phosphatase. Cancer Res. 2009;69:7739–7746. doi: 10.1158/0008-5472.CAN-09-1805. [DOI] [PubMed] [Google Scholar]
- 54.Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581:3545–3549. doi: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- 55.Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, Taylor SS. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J. Biol. Chem. 2011;286:2918–2932. doi: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.