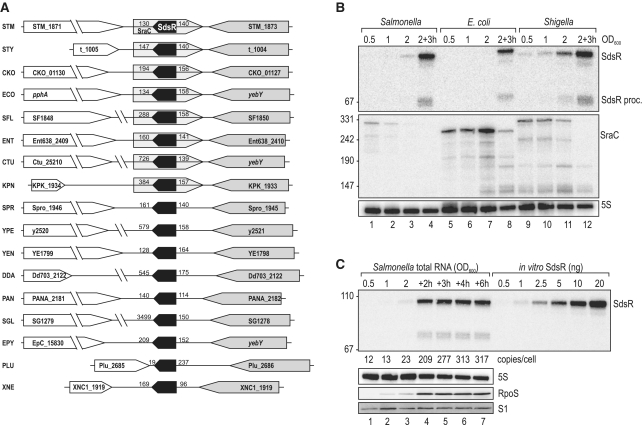
Figure 1.
(A) Genomic context of the sdsR gene in various Enterobacteria. Synteny analysis of the sdsR/sraC genes in various enterobacterial species revealed partial conservation of the locus. In all cases sdsR is positioned downstream of yebY or homologues; the flanking gene at the 3′-end is variable. Distances to flanking genes are indicated in bp. STM: Salmonella typhimurium; STY: Salmonella typhi; CKO: Citrobacter koseri; ECO: Escherichia coli; SFL: Shigella flexneri; ENT: Enterobacter; CTU: Cronobacter turicensis; KPN: Klebsiella pneumoniae; SPR: Serratia proteamaculans; YPE: Yersinia pestis; YEN: Yersinia enterolitica; DDA: Dickeya dadantii; PAN: Pantoea ananatis; SGL: Sodalis glossinidius; EPY: Erwinia pyrifoliae; PLU: Photorhabdus luminescens; XNE: Xenorhabdus nematophila. (B) Expression levels of SdsR and SraC sRNAs in Salmonella, E. coli and Shigella. Northern blot analysis of total RNA isolated from wild-type Salmonella (JVS-1574), E. coli (JVS-5105) and Shigella (JVS-0012) cells grown to OD600 of 0.5, 1.0, 2.0 and 3 h after cells had reached an OD600 of 2.0. SdsR and SraC sRNAs were detected by radio-labelled oligo probes directed against the conserved sRNA sequences. 5S rRNA levels were determined to confirm equal loading. (C) SdsR copy number over growth. SdsR levels at various timepoints (OD600 of 0.5, 1.0, 2.0 and 2, 3, 4 or 6 h after cells had reached an OD600 of 2.0) in wild-type Salmonella were compared by northern blotting to signals of in vitro transcribed SdsR in indicated amounts. SdsR was detected using a riboprobe; probing for 5S RNA confirmed equal loading. Expression of RpoS at different growth stages was determined by western blotting. Detection of ribosomal protein S1 was used as loading control.