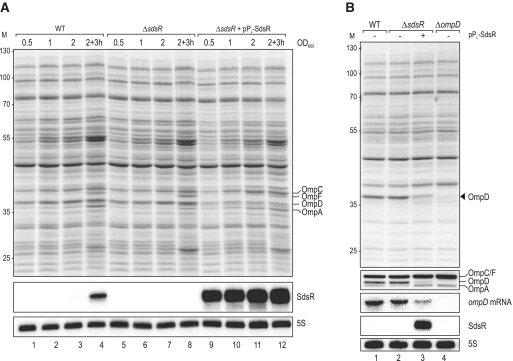
Figure 3.
(A) Proteome changes upon SdsR over-expression in Salmonella. Whole-cell protein patterns of wild-type and ΔsdsR Salmonella carrying either a control vector (pJV300) or the constitutive SdsR-expression plasmid pPL-SdsR (pKF68-3) grown in LB were compared by separation of total cell lysates from several conditions (OD600 of 0.5 (lanes 1, 5, 9); 1.0 (lanes 2, 6, 10); 2.0 (lanes 3, 6, 11); 3 h after cells had reached an OD600 of 2.0 (lanes 4, 9, 14)) by 11% SDS–PAGE; the gel was stained for abundant proteins with Coomassie Blue. Sizes of co-migrating marker proteins are marked at the left in kDa. Positions of the major porins OmpC, OmpF, OmpD and OmpA are indicated. SdsR expression was determined by northern blotting of RNA isolated from the same cultures and loading was controlled by probing for 5S RNA. (B) Verification of specific OmpD repression by SdsR. Wild-type, ΔsdsR and ΔompD (JVS-0735) Salmonella transformed with either the control vector or pPL-SdsR were grown to an OD600 of 2.0, and OmpD protein levels were analysed by SDS-PAGE (top panel) and western blotting using an antiserum detecting the major Salmonella porins as indicated (second panel). Northern blot analysis (three lower panels) of the same strains revealed reduced ompD mRNA steady-state levels in cells over-expressing SdsR. 5S RNA served as loading control.