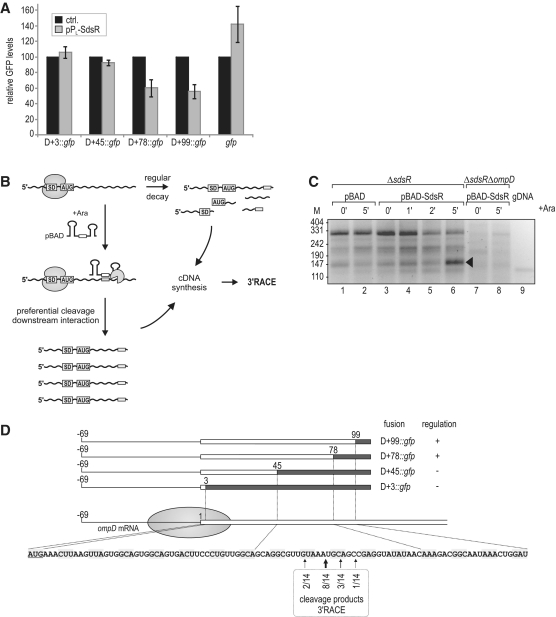
Figure 5.
(A) Regulation of OmpD-GFP reporter fusions by SdsR. Salmonella ΔsdsR ΔompD cells carrying the control vector or pPL-SdsR were co-transformed with low-copy plasmids expressing gfp alone or a series of translational ompD::gfp fusions spanning the complete 5′-UTR plus an increasing number of nucleotides of the ompD coding sequence (D+3::gfp; D+45::gfp; D+78::gfp; D+99::gfp; see Supplementary Table S2 for details on plasmids) as depicted in (D). Whole-protein samples were collected from cells grown to an OD600 of 2.0, and regulation of reporter fusions was determined by signal quantification on western blots. Relative GFP levels in the presence of the control plasmid (black bars; set to 100) or the constitutive pPL-SdsR (grey bars); errors indicate standard deviation from three biological replicates. (B) Schematic illustration of the 3′-RACE approach employed for target site determination. (C) 3′-RACE analysis of ompD mRNA fragments enriched upon SdsR pulse expression. cDNA was prepared from total RNA of ΔsdsR cells as well as the ΔsdsR ΔompD control strain prior to and at indicated timepoints after SdsR induction from an inducible PBAD promoter. Salmonella genomic DNA (gDNA) served as a control template. DNA fragments were recovered from the indicated band of ∼150 bp (lane 6), and ompD 3′-ends were determined by sequencing of subcloned fragments. (D) Location of ompD 3′-ends obtained by 3′-RACE analysis. The ompD::gfp reporter plasmids and their regulation by SdsR (see Figure 5A) are represented schematically. The filled circle indicates the approximate coverage of ompD mRNA by the 30S ribosomal subunit binding to the RBS. Position as well as frequency of enriched break-down products determined by 3′-RACE (Figure 5C) are shown below the ompD CDS.