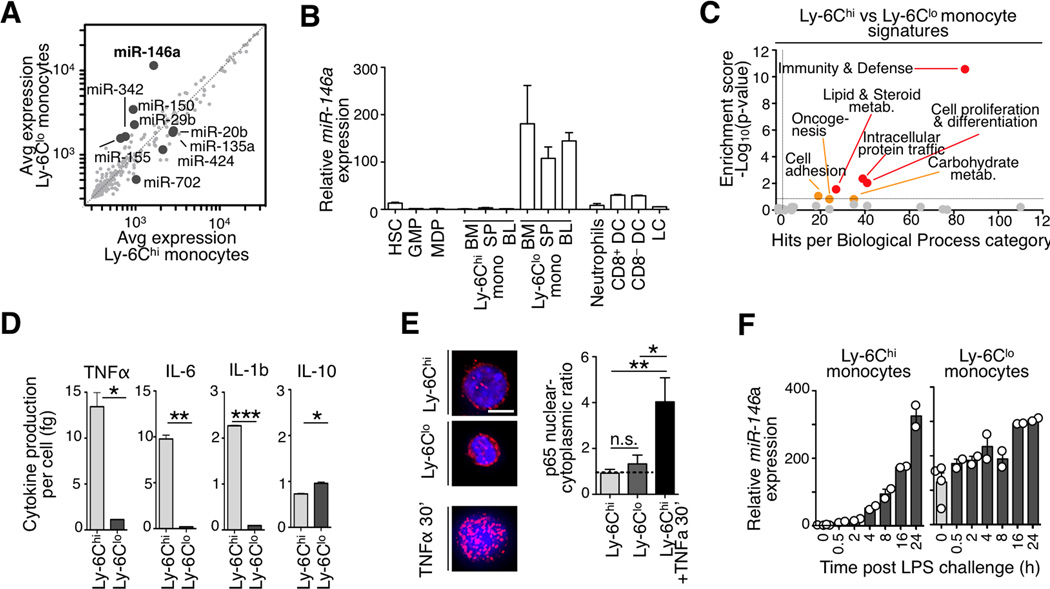
Figure 1. Mouse monocyte subsets show distinct miR-146a and inflammatory profiles.
a) Microarray analysis of miRNA expression in Ly-6Chi vs. Ly-6Clo splenic monocytes. Genes with >2-fold change among subsets and p<0.05 are highlighted (n=4 biological replicates).
b) Relative miR-146a expression in various hematopoietic cell types. Expression is relative to splenic Ly-6Chi monocytes (n=3 animals for all cell populations except for spleen monocytes, n=7).
c) Analysis of differentially expressed genes in Ly-6Chi vs. Ly-6Clo blood monocytes using the Panther database of biological functional categories.
d) Quantification of TNFα, IL-6, IL-1b and IL-10 production by splenic monocytes 8 h after LPS challenge (n=3–4).
e) p65 immunofluoresence staining in sorted monocyte subsets. Ly-6Chi monocytes stimulated for 30’ with TNFα prior to fixation served as a positive control for effective nuclear translocation. Scale bar: 10 µm
f) Time-course analysis of miR-146a levels after LPS challenge. Expression is relative to Ly-6Chi monocytes at time 0 h (n=2–4).
Data are presented as mean±SEM. (* p<0.05,** p<0.01, *** p<0.001, Student’s t-test).