Abstract
A recent study using massive parallel sequencing demonstrates unequivocally that alternative tandem acceptor splicing is tissue-specifically regulated.
Keywords: Alternative splicing, bioinformatics, phylogenetic conservation, RNA-seq, splicing mechanisms, splicing regulation, tandem splice acceptors
Different protein isoforms are produced from a single gene through alternative splicing. The inclusion or exclusion of an exon, also referred to as exon skipping, is the most prevalent form of alternative splicing, followed by the usage of alternative 3' (acceptor) or 5' (donor) splice sites. Often these alternative splice sites can be found in very close proximity to each other. Alternative acceptor motifs 3 bp apart from each other, also referred to as NAGNAG or tandem acceptor sites, can be found in 30% of human genes and appear to be functional in at least 5% [1] (Figure 1, upper panel). Their alternative usage alters the transcript by the length of exactly one codon and, thus, is reading frame preserving. Among all reading-frame-preserving alternative splicing events, NAGNAG splicing is the second most frequent form (approximately 20%) after exon skipping (approximately 60%) [2]. The two alternative mRNA isoforms differ in length by three nucleotides, altering one or two amino acids in the resulting protein depending on the phase of the intron [1]. The upstream NAG is referred to as the proximal acceptor and the downstream NAG is referred to as the distal acceptor. A recent study by Bradley et al. [2] presents the first genome-wide study analyzing the potential regulation of NAGNAG alternative splicing using massive parallel sequencing.
Figure 1.
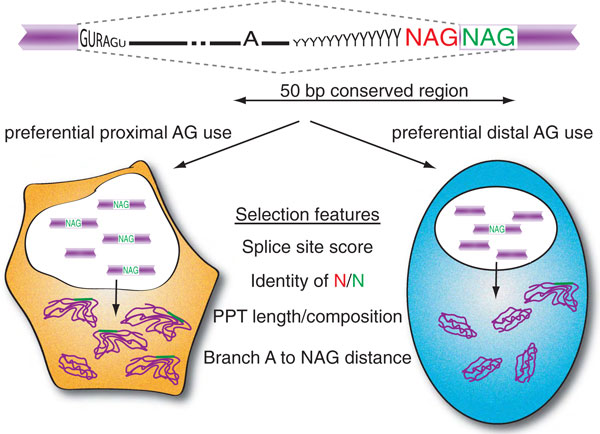
Tissue-specific alternative splicing at NAGNAG splice sites. Many human exons contain tandem acceptor sites (NAGNAG) that are often subject to alternative splicing. Massive parallel sequencing of tissue-derived transcriptomes now proves that this type of alternative splicing is regulated [2]. The proximal (red) and distal (green) splice sites compete for splice site pairing. Tissue-specific splicing preferences (left and right pathways) depend on highly conserved intronic sequences within 50 bp upstream of the NAGNAG motif. The most likely features that mediate alternative NAGNAG splicing are the strength of the splice sites, the identity of the competing NAGs, the polypyrimidine tract (PPT) length and composition, and the distance between the branch point sequence and the NAGNAG motif.
Since the discovery of NAGNAG alternative splicing almost a decade ago, debates about its functional implications and regulatory mechanisms have been controversial. Several lines of work focused on delineating whether alternative acceptor site selection is actively regulated or stochastically selected during the splicing process. For example, it was argued that constitutive or alternative usage of tandem acceptors strictly depends on the immediate sequence context around NAGNAG sites, thus proposing a simple physical model that implies a stochastic selection process [3]. Similarly, machine learning approaches based on such sequence features proved very reliable in predicting whether a NAGNAG configuration at a 3' splice site would elicit alternative splicing [4]. Other approaches supplied indications of regulation. Analyzing splicing events covered by a large number of EST entries suggested that some alternative NAGNAG splicing may be tissue specific [1], a proposal supported by high evolutionary conservation and an overabundance of cis-regulatory elements in proximity of alternatively spliced NAGNAGs [5]. Such regulation would imply a biological function to alternative NAGNAG splicing rather then just an imprecision of the spliceosome. The notion of functionally important alternative NAGNAG splicing was further promoted by the demonstration that such events are selected against within parts of an ordered three-dimensional protein structure [6]. However, even with all these interesting puzzle pieces provided over the last years, it was still not clear if alternative NAGNAG splicing is the result of yet another form of regulated mRNA diversification, or whether it is simply a reflection of splicing inaccuracies that appear to be particularly abundant when splice junctions are presented in such close proximity. While several experimental attempts have been made to identify examples of regulated NAGNAG alternative splicing, most datasets analyzed were so limited in number that no convincing general conclusions could be drawn. Thus, the jury was still out to characterize NAGNAG alternative splicing.
In a recent issue of PLoS Biology, Bradley et al. [2] present the first genome-wide study analyzing the potential regulation of NAGNAG alternative splicing using RNA-Seq data from 16 human tissues as well as 8 mouse tissues. This 'tour de force' dataset allowed Bradley et al. to first identify alternatively used NAGNAG motifs and then ask how many of them display altered usage in the various tissues analyzed. The answers from this study are clear and undisputable. Of the more than 2,000 alternatively used NAGNAG motifs in human protein-coding regions, 73% showed evidence for tissue-specific regulation, with more than 40% displaying major changes in the proximal to distal ratio (>25% change between tissues). Thus, most alternative NAGNAG splice sites are subject to some form of regulation. Moreover, Bradley et al. found a positive correlation between the magnitude of tissue-specific splicing differences and the conservation of the NAGNAG alternative event, suggesting that regulated NAGNAG alternative splicing has been evolutionarily fixed to retain an advantageous function for the cell.
Having demonstrated that indeed NAGNAG alternative splicing displays tissue-specific splicing patterns, Bradley et al. tackled the next phase of investigation: deciphering potential mechanisms that mediate these observed changes in proximal and distal splice site usage. The reason why this line of investigation is of particular interest is the fact that the close proximity between the alternative splice sites prohibits the assembly of the typical splicing regulatory factors that tend to bind between competing alternative splice sites to direct splice site selection. For NAGNAG alternative splicing such mechanisms are considered practically impossible because the association of splicing regulators within the pre-mRNA requires a binding platform of at least five or six nucleotides. Furthermore, it is unclear at which step during spliceosomal assembly and catalysis alternative AG selection is set in stone. Given the observation that the first step of splicing can occur even in the absence of a NAG intron/exon junction [7], it is likely that alternative NAGNAG choice could occur during the second step of the splicing reaction, after major structural rearrangements of the spliceosome. If this is indeed the case, it is possible that the mechanisms modulating tissue-specific alternative NAGNAG usage might conceptually be different from the already established mechanisms of alternative splicing.
Relying on phylogenetic conservation, Bradley et al. argue that a 50 bp region upstream of tissue-specific NAGNAGs contains most potential regulatory information responsible for alternative splicing [2] (Figure 1). In agreement with previous work [3,4], Bradley et al. suggest a biophysical model to predict the basic quantitative NAGNAG isoform usage by simply applying the splice site scores of the competing alternative acceptors. In addition, the length and composition of the polypyrimidine tract appears to correlate with the control of NAGNAG splicing, as does the distance between the splice site and the branch point sequence, observations that confirm earlier reports [8,9]. These results are reminiscent of studies that demonstrated that preferential distal AG selection depends on the distance between the branch point sequence and the proximal AG to avoid the usual selection of the proximal splice site [10]. All in all, these analyses indicate that regulated NAGNAG splicing events are classified by a particular set of architectural features within the 3' splice site region, presumably to promote differential AG selection at the variable conditions unique to the tissues investigated.
However, even with these findings we can only speculate how NAGNAG splicing events are regulated in a tissue-specific manner. Common sense presumes the existence of binding sites for regulatory factors, which themselves might show tissue-specific expression. However, neither the sequence feature analysis nor the physical requirement for binding-site real estate points at this stage towards any of the usual suspects (serine/arginine (SR)-rich proteins and heterogeneous nuclear ribonucleoprotein (hnRNP) proteins that have been associated with regulated alternative splicing). One of the critical issues to solve is to determine when during the splicing reaction alternative AG selection is carried out. Does it occur during initial splice site selection as is common for most other types of alternative splicing, or is alternative NAGNAG selection set in stone during the second step of the splicing reaction as the spliceosome rearranges its active site, as we speculated earlier? Identifying the step(s) associated with alternative NAGNAG splicing will be likely to open the door for more directed experimental and computational approaches to figure out the mechanisms behind these indel alternative splicing events. Along the road to discovery it might be wise to equally concentrate on the minor fraction of NAGNAG motifs that do not display regulated alternative splicing. Given their smaller number, they may hold the key to elucidate the mechanisms.
It is fair to say that the evidence provided by the work of Bradley et al. should bring the discussion of whether alternative NAGNAG splicing is regulated or stochastic to an end [2]. The realization of extensive NAGNAG splicing differences across various tissues will challenge new investigations to determine how regulation is achieved. While it is possible that not all alternative NAGNAG events are controlled similarly, future studies will be likely to uncover new mechanisms to guide the selection of close proximity 3' splice sites. Based on the discoveries made by Bradley et al., we may be able to look forward to yet other interesting forms of splice pattern regulation.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Anke Busch, Email: abusch@uci.edu.
Klemens J Hertel, Email: khertel@uci.edu.
Acknowledgements
This authors are supported by grants from the NIH (RO1 GM62287 and R21 CA149548 to KJH) and from a fellowship within the Postdoc Programme of the German Academic Exchange Service, DAAD (AB).
References
- Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat Genet. 2004;36:1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
- Bradley RK, Merkin J, Lambert NJ, Burge CB. Alternative splicing of RNA triplets is often regulated and accelerates proteome evolution. PLoS Biol. 2012;10:e1001229. doi: 10.1371/journal.pbio.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern TM, van Nimwegen E, Kai C, Kawai J, Carninci P, Hayashizaki Y, Zavolan M. A simple physical model predicts small exon length variations. PLoS Genet. 2006;2:e45. doi: 10.1371/journal.pgen.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Nikolajewa S, Szafranski K, Hiller M, Jahn N, Huse K, Platzer M, Backofen R. Accurate prediction of NAGNAG alternative splicing. Nucleic Acids Res. 2009;37:3569–3579. doi: 10.1093/nar/gkp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman M, Mandel-Gutfreund Y. Alternative splicing regulation at tandem 3' splice sites. Nucleic Acids Res. 2006;34:23–31. doi: 10.1093/nar/gkj408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Szafranski K, Huse K, Backofen R, Platzer M. Selection against tandem splice sites affecting structured protein regions. BMC Evol Biol. 2008;8:89. doi: 10.1186/1471-2148-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K, Moore MJ. Bimolecular exon ligation by the human spliceosome. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- Tsai KW, Chan WC, Hsu CN, Lin WC. Sequence features involved in the mechanism of 3' splice junction wobbling. BMC Mol Biol. 2010;11:34. doi: 10.1186/1471-2199-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KW, Tarn WY, Lin WC. Wobble splicing reveals the role of the branch point sequence-to-NAGNAG region in 3' tandem splice site selection. Mol Cell Biol. 2007;27:5835–5848. doi: 10.1128/MCB.00363-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K, Reed R. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol Cell Biol. 2001;21:1509–1514. doi: 10.1128/MCB.21.5.1509-1514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]