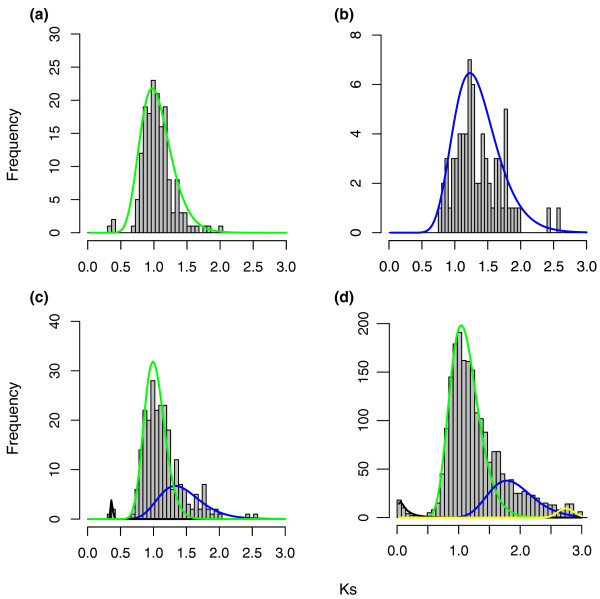
Figure 5.
Ks distributions of paralogs in Vitis from syntenic block analysis. Methods for sequence alignment and estimation of Ks were as reported (Cui et al. 2006), but were here limited to paralogous gene pairs retained on syntenic blocks in the Vitis genome. Colored lines superimposed on Ks distribution represent significant duplication components identified by likelihood mixture model as in Figure 4 (Materials and methods). a, Ks distribution of 168 Vitis pairs supporting core eudicot-wide duplication in phylogenetic analysis. One statistically significant component: green/1.03/1. b, Ks distribution of 70 Vitis pairs showing all eudicot-wide duplications on phylogenies. One significant component: blue/1.31/1. c, Ks distribution of combination of Vitis pairs supporting core eudicot- (a) and eudicot-wide duplications (b) on phylogenies. Three significant components: black/0.3/0.01, green/1.02/0.70, blue/1.40/0.29. d, Ks distribution of 2191 paralogous pairs were identified from syntenic block analysis. Four significant components: black/0.12/0.02, green/1.09/0.74, blue/1.85/0.22, yellow/2.7/0.02.