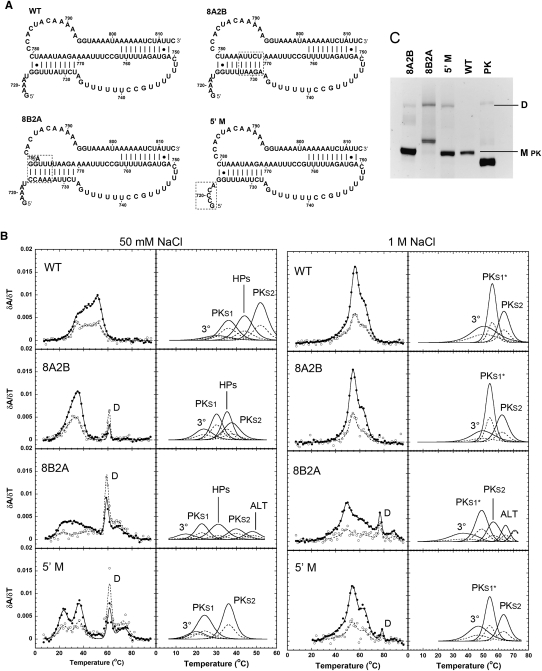
FIGURE 5.
Comparisons between the WT (WT) pseudoknot domain construct and stem 1 and 5′-terminal mutant RNAs. (A) RNA constructs for the wild-type pseudoknot region (WT) and the 8A2B, 8B2A, and Δ5′ mutations. Dashed boxes enclose the mutated regions. (B) Profiles on the left show every fifth data point obtained at 260 nm (●) and 280 nm (○), and the calculated fits to the 260-nm (solid lines) and 280-nm data (dashed lines), with the individual transitions at 260 nm (solid lines) and 280 nm (dashed lines), which combine to generate the total calculated curves, shown separately to the right. The individual transition fits are labeled according to the correlated structural elements: (3°) tertiary structure, (PKS1) pseudoknot stem 1, (HPs) the overlapped H5 and S2* transitions, (PKS1*) pseudoknot stem 1 and 6-bp helix, (PKS2) pseudoknot stem 2, (ALT) alternative structure, respectively. (C) Native polyacrylamide gel. Monomer (M pk) and dimer (D) species are indicated to the right of the gel, and the PK RNA was loaded for a size comparison to the monomeric species.