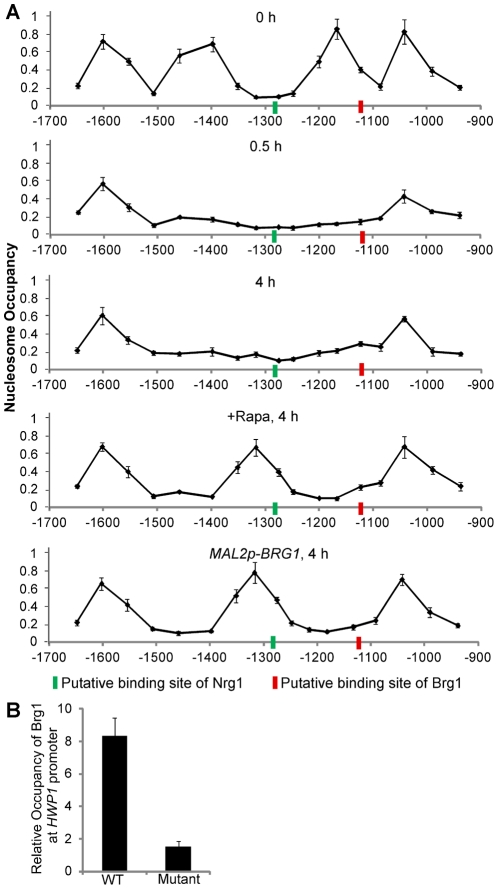
Figure 5. Nucleosome mapping analysis at the UAS region of the HWP1 promoter indicates different promoter accessibility to Nrg1 and Brg1 in yeast and hyphal cells.
Overnight cultures of wild type (SN250) cells were diluted 1∶100 fold into YPD medium in the presence or absence of 10 nM rapamycin at 37°C. Cells of wild type carrying MAL2p-BRG1 (HLY3636) were diluted 1∶100 fold into YPMaltose medium at 37°C. Cells were collected at indicated times and conditions for nucleosome mapping analysis. PCR primer pairs were designed to generate ∼100 bp fragments, with ∼60 bp overlapping and a 40 bp gap between neighboring PCR fragments. The x-axis represents the midpoints of qPCR reactions. The signals from genomic DNA from each sample for each primer pair were used for normalization. Data show the average of three independent qPCR experiments with error bars representing the SEM. Putative binding sites of Nrg1 (−1284∼−1279) and Brg1 (−1130∼−1125) in the UAS region of the HWP1 promoter are marked. (B). The predicted Brg1 binding motif in the UAS region of the HWP1 promoter is required for Brg1 binding. An overnight culture of HLY4079 (carrying both the WT HWP1 promoter and a copy of the HWP1 promoter mutated at the Brg1 binding site) was inoculated at a 1∶100 dilution into YPD medium in the presence of 10 nM rapamycin at 37°C for 4 h. Primers 21 and 22 were used to quantitate the binding of Brg1 to the WT HWP1 promoter over no tag control, and primers 21 and 23 were used to quantitate the binding of Brg1 to the mutated HWP1 promoter over no tag control.